Abstract
Background:
Hirschsprung disease (HSCR) is the third most common congenital disorder of the gastrointestinal tract. This study aims to elucidate changes in protein expression between the normal and aganglionic hindgut in human HSCR.
Methods:
The biopsies were obtained from the normal and aganglionic hindgut in human HSCR, and the comparative proteomics were analyzed by mass spectrometry (MS)–based two-dimensional gel electrophoresis (2DE).
Results:
A total of 932–986 protein spots were identified in each of the gut segments, among which 30 spots had at least an eightfold difference in volume (%). Of the 30 differentially expressed spots, 15 proteins were identified via sequence analysis. Among these 15 proteins, eight were upregulated and seven were downregulated in the aganglionic group. The well-represented classes included biomarkers of enteric ganglions, extracellular matrix proteins, LIM domain proteins, serum proteins, and other pleiotropic proteins. Five proteins were selected and verified by western blotting and real-time PCR, and the results were consistent with the results of 2DE.
Conclusion:
MS-based 2DE can help to identify pathological relevant proteins in HSCR; it defines an extensive protein catalog of the normal and aganglionic hindgut and may constitute the basis to understand pathophysiological mechanisms related to the HSCR.
Similar content being viewed by others
Main
Hirschsprung disease (HSCR) is one of the most common congenital malformations of enteric nervous system (ENS) in children. It is the third most common congenital disorder of the gastrointestinal tract worldwide, and it occurs in 1/5,000 live births (1,2,3). HSCR is characterized by the absence of ganglion cells in the myenteric and submucosal plexuses of the gastrointestinal tract, resulting in intestinal obstruction and constipation in neonates and children (2). It is caused by an anomalous ENS and is therefore considered to be a neurocristopathy. The ENS is formed from a multipotent progenitor cell population known as the enteric neural crest cells (ENCCs), which are derived from the neuroepithelium. ENCCs migrate over substantial distances to colonize the entire length of the gut, and during their migration, they need to survive, proliferate, and ultimately differentiate. The absence of an ENS from variable lengths of the colon results in HSCR or aganglionosis (3,4,5).
It is well established that HSCR is a set of complex diseases with extensive molecular genetics bases. HSCR is caused by several factors, and at least 10 different genes and 5 chromosomal loci contribute to its pathogenesis (4,5,6).Among the genes with the highest expression levels were GDNF, Sox10, GFRα1, and EDNRB. It has been indicated that mutations in these genes lead to long-segment HSCR and syndromic HSCR. Other differentially expressed genes belonging to several different functional categories were also identified. Among the well-represented classes were transcription factors (e.g., Hox, Sox, T-box, and Ets family transcription factors), genes involved in cell adhesion (e.g., cadherins, protocadherins, and cell adhesion molecules), and microtubule-associated proteins (e.g., stathmin family proteins) (7,8). Mediators of several well-described signaling pathways, including receptor ligands (e.g., fibroblast growth factors and chemokines), kinases, and G-protein subunits, were also identified (8). All the genes were expressed in the embryonic murine gut and in the regions of ENS formation, validating the results of the microarray analyses (7,8). Hence, HSCR is considered to be a genetic hereditary disease in most cases.
However, mRNA levels do not always correspond to protein expression or activity. This is due to several posttranslational modifications such as ubiquitination, protease-mediated cleavage, glycosylation, phosphorylation, methylation, and acetylation (9). As a result, HSCR has been considered not only genetically but also clinically abnormal. Proteomic approaches can present advantages over genomic approaches because protein biomarkers are the direct functional players compared with transcripts (10,11). Standard proteomic techniques, including two-dimensional gel electrophoresis (2DE) and mass spectrometry (MS), have been under development for the past three decades, beginning at the end of the 1990s. Due to the development of high-throughput platforms, proteomic analyses are no longer limited to the examination of a few select proteins at a given time, and the simultaneous measurement of multiple protein products and/or protein modifications can now be performed (12,13). Moreover, such analyses allow for the comparison of protein expression in different samples, facilitating the comparison of their proteomic profiles to identify the differentially expressed proteins that are associated with multiple diseases, such as cancer and neurocristopathy (12,13,14).
In this study, MS-based 2DE was applied to study samples of normal and aganglionic segments from humans with HSCR to detect the proteins that are relevant to the development of this disease.
Results
Protein Profiles of the Normal and Aganglionic Hindgut
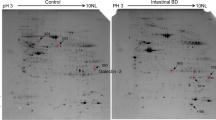
The proteomic analysis of the normal colon and the ganglionic and aganglionic hindgut samples revealed 932–986 detectable spots on a single modified colloidal Coomassie Brilliant Blue (mcCBB)-stained 2DE gel ( Figure 1 ). More than 80% of the protein spots were consistent between the three sets of CBB-stained gels. Overall, the pattern of 2DE protein spots across the gels was similar for the normal, ganglionic, and aganglionic hindgut samples. Between the tissues from normal and aganglionic hindguts, at least a twofold difference in volume (%) was observed for 158 protein spots (P < 0.05), among which 30 protein spots were found to exhibit at least an eightfold difference in volume (%) (P < 0.05). These 30 protein spots were dissected for further analysis, and their expression levels were the same in the normal and ganglionic hindguts. Of the 30 differentially expressed spots, 15 proteins were identified based on MS. Among these 15 proteins, eight were upregulated and seven were downregulated ( Table 1 ).
Two-dimensional gel electrophoresis (2DE) analysis of the proteomic profiles of the normal, ganglionic, and aganglionic hindgut. Representative Coomassie-stained 2DE gels showing the protein expression maps in the hindgut from (a) normal colon tissue from a patient with trauma to the colon (normal group), (b) a ganglionic segment from a Hirschsprung disease (HSCR) patient (ganglionic group), and (c) an aganglionic segment from a HSCR patient (aganglionic group). The numbers indicate the differently expressed protein spots that are presented in Table 2 . Accession numbers are indicated.
After carefully analyzing the structure and function of these proteins, several proteomic candidates with different biological characteristics were identified ( Table 2 ). We selected five proteins, sox10, fibrinogen (Fn), isoform 1 of four and a half LIM domains protein 1 (FHL1), transgelin (TAGLN), and anterior gradient protein 2 homolog (AGR2), for further confirmation.
Western Blotting
The expression of the selected proteins was quantitatively studied by performing western blot analysis. The expression of FHL1, TAGLN, and AGR2 was upregulated 2.0-, 5.3-, and 4.8-fold, respectively, in the aganglionic segment compared with their expression in the normal group (P < 0.05, Figure 2b , c ), whereas Fn was downregulated by 60%, and Sox10 was not detected in these samples (P < 0.05, Figure 2b , c ). The results were consistent with what was observed in the 2DE ( Figure 2a and Table 1 ). Importantly, the expression of these proteins did not differ significantly in the samples that were obtained from the normal and ganglionic segments (P > 0.05).
Western blot analysis of selected proteins. (a) Confirmation of two-dimensional gel electrophoresis results by western blotting. Analysis of the expression of selected proteins using actin as an internal control. (b) Quantification of the western blot results. Four and a half LIM domains protein 1 (FHL1), transgelin (TAGLN), and anterior gradient protein 2 homolog (AGR2) were upregulated 2.0-, 5.3-, and 4.8-fold, respectively, in aganglionic segments, whereas Fn was downregulated 20%, and Sox10 was not detected in these samples. *P < 0.05. No fill = normal (n = 4), gray fill = ganglionic segment (n = 6), and black fill = aganglionic segment (n = 6).
Real-Time Quantitative PCR
The mRNA expression of the selected proteins was studied by real-time quantitative PCR analysis. The mRNA levels of FHL1, TAGLN, and AGR2 were upregulated 2.6-, 5.8-, and 4.2-fold, respectively, in the aganglionic segment compared with their levels in the normal group (P < 0.05), whereas the mRNA levels of Fn and Sox10 were downregulated by 26 and 10%, respectively, in these samples (P < 0.05, Figure 3 ). These results were consistent with the results of the 2DE and western blotting. The samples from the normal and ganglionic segments did not differ significantly in terms of mRNA expression (P > 0.05, Figure 3 ).
Real-time quantitative PCR analysis of the selected proteins. The mRNA levels of four and a half LIM domains protein 1 (FHL1), transgelin (TAGLN), and anterior gradient protein 2 homolog (AGR2) were upregulated 2.6-, 5.8-, and 4.2-fold, respectively, in aganglionic segments, whereas the mRNA levels of Fn and Sox10 were downregulated 26 and 10%, respectively, in these samples. *P < 0.05. No fill = normal (n = 4), gray fill = ganglionic segment (n = 6), and black fill = aganglionic segment (n = 6).
Discussion
HSCR is an anomaly of ENS, which is characterized by the absence of myenteric and submucosal plexus in the bowel. Recent advances in proteomic analysis and bioinformatics have revolutionized our understanding of the underlying mechanisms of HSCR (15,16,17). Using an MS-based 2DE technique, the Hagl and Marvin-Guy groups conducted proteomic analyses of myenteric plexus in rat gut, and they identified several novel protein biomarkers that were associated with ENS development (15,16). Both of these studies were conducted using ENCCs and could therefore only reflect the roles of ENCCs. But it has been known that the roles of the enteric microenvironment, which interacts with ENCCs, appear to be more critical in this condition. Taking this into consideration, Gao et al. (17) conducted a proteomic analysis of the colonic tissues from HSCR patients using the same technique. A few valuable protein biomarkers associated with HSCR were enriched, but the scope of the results was limited. Herein, we observed the differential protein expression in paired colonic tissues from HSCR patients. The obtained data suggest that the proteomic profiles are significantly different between the normal and aganglionic hindgut in HSCR, and several candidate markers that are associated with different biological activities were identified.
Sox10 and hnRNPA2/B1
Sox10 is a member of the SRY-box-containing family of transcription factors (18,19). It has been demonstrated that Sox10 promotes the proliferation and survival of ENCCs, and it is critical for the maintenance of the ENS progenitor pool (19). Other studies have implicated that Sox10 can regulate the expression of c-Ret and Phox2b (20). In mice, the roles of Sox10 in HSCR, Waardenburg syndrome type 2 and type 4, and neural crest development have been extensively described. The present proteomic study indicated that the Sox10 protein was absent and that the Sox10 mRNA levels were low in the aganglionic hindguts from human HSCR, which is consistent with the results of previous studies (19,20). Heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNP A2/B1) is a member of the hnRNP family of proteins. The hnRNPs are a complex group of RNA-binding proteins that play a key role in mRNA processing and telomere biogenesis (21,22,23,24,25). They bind to nascent RNA polymerase transcripts and are involved in various aspects of pre-mRNA processing, mRNA transport, mRNA metabolism, and alternative splicing (21,22). There is increasing evidence of a prominent role for hnRNP A2/B1 in the regulation of cell metabolism, migration and invasion, proliferation, and cellular response to mitochondrial stress. However, there is no described mechanistic link for the contribution of hnRNP A2/B1 overexpression and the fate of ENCCs; therefore, we cannot determine their roles in ENS formation or HSCR histogenesis, although a significantly higher protein level of hnRNP A2/B1 has been detected in the present proteomic study. We were unable to ascertain whether these two markers are either the causes or the effects of neuronal absence in HSCR because they are both enteric neuron-related biomarkers. However, we have reason to believe that these markers could be used as valuable neuronal biomarkers for the diagnosis or differentiation of HSCR.
Fn, LAMA3, and SERPINA1
Based on their structure and biological function, Fn, LAMA3, and SERPINA1 were all found to be extracellular matrix (ECM) proteins. ECM is a complex mixture of matrix molecules, including the glycoproteins fibronectin, collagens, laminins, proteoglycans, and nonmatrix proteins. ECM induces discrete cell surface structures that are tightly associated with the matrix that are termed cell–matrix adhesions, and it regulates biological processes, such as cell survival, cell proliferation, wound healing, and tumorigenesis (26). The roles of ECM in ENS have been well described that abnormal ECM components or improper extracellular signals could lead to various ENS diseases, such as HSCR (27). As the major components of ECM, Fn and LAMA3 have both been renowned for their confirmed functions during ENS morphogenesis (26,27). It has also been reported that Fn is abnormally expressed in the aganglionic segment of patients with HSCR, resulting in the faulty migration of ENCCs and in the pathogenesis of aganglionosis or HSCR (28). Laminin is a cross-shaped heterotrimeric protein composed of three chains (29). It has been suggested that laminin expression changes in the gut environment, which occurs normally during development and creates a physical obstacle to a delayed wavefront of ENCCs, as was reported for EDNRB mutant ENCCs by these authors (29,30). In the present study, both Fn and LAMA3 proteins were indicated to be abnormally expressed in the aganglionic hindguts from human HSCR, the results were consistent with those of previous studies (29,30). In addition to Fn and laminin, another ECM protein, SERPINA1, was also detected using the present proteomic study. SERPINA1 is a proteoglycan that is synthesized by the gut. The roles of SERPINA1 in HSCR have not been well explored and require further study.
FHL1, LASP1, and TES
Based on their structural analysis, these three proteins are found to contain similar structural domains—LIM domains. The LIM domains contain eight conserved residues—primarily cysteines and histidines—that coordinately bind to two zinc atoms and are named after their initial discovery in the proteins Lin11, Isl-1, and Mec-3. They frequently occur in multiples, as seen in proteins, such as TES and LMO4, and they can also be attached to other domains to confer a binding or targeting function upon them, such as LIM-kinase. With the increasing number and functional diversity of LIM domain proteins, they are further classified into many groups with diverse biological functions based on their molecular structures (31). LIM domain proteins can mediate protein:protein interactions that are critical to cellular processes. Through the binding of their partners, LIM proteins play critical roles in the regulation of actin structure and dynamics, neuronal pathways, integrin-dependent adhesion and signaling, cell fate determination, and tissue-specific gene expression (31,32). As members of different LIM protein groups, three LIM domain proteins, FHL1, LASP1, and TES, were identified in this study. These proteins have been proven to act as linkers that connect cell-ECM adhesions to the actin cytoskeleton, and they participate in the assembly of cell-ECM attachments, cell proliferation, and cell migration (33,34,35). However, at present, there is no direct evidence for the contribution of these LIM proteins in deciding the fate of ENCCs. Although we cannot conclude their roles in ENS formation or HSCR histogenesis, significantly differential expression levels have been observed in these proteins. The study of LIM domain-containing proteins in ENCC proliferation and migration is a promising field in HSCR histogenesis. These studies are being carried out in our laboratory, and they will be presented in the near future.
Other Pleiotropic Proteins
The remaining proteins that were identified in the present study include the serum proteins and other pleiotropic proteins. The serum biomarkers that are associated with HSCR have been comprehensively studied by another group and are not discussed here (36). The pleiotropic proteins TAGLN, CLCA1, AGR2, and HSPB1 are a group of proteins with diverse functions, but they could not be classified into a special subtype. TAGLN is a member of the calponin family of actin-binding proteins, and it is localized to the cytoskeletal apparatus (37). TAGLN has been reported to be involved in cell migration because of its roles in the formation of actin filament bundles that form podosomes and in suppressing the expression of metallo-matrix proteinase-9 (MMP-9), which is involved in tissue remodeling and cell adhesions. In addition, TAGLN has been hypothesized to regulate the contractile function of smooth muscle cells (37). In the present study, TAGLN protein was found to be upregulated in the aganglionic hindgut, indicating that it plays a role in this disease. The role of TAGLN in HSCR has not been determined, although its contribution to the motility dysfunction of the aganglionic hindgut is tenable. AGR2 is another upregulated protein in the present study, and it was proven to be expressed in the surface epithelial and submucosal glands. Its localization allows for it to serve as a regulator of enteric content. AGR2 was recently classified as a member of the protein disulfide isomerase family, and it was demonstrated to play a special role in the production of mucus (38). AGR2-deficient mice cannot produce intestinal mucin and are highly susceptible to experimentally induced colitis. These effects can explain the phenomenon that the aganglionic segment in HSCR is more prone to result in enterocolitis. Based on these findings, we propose a role for AGR2 in HSCR-related enterocolitis, but further studies are required for confirmation.
Our proteomics analysis suggests a role for the perturbation of the protein folding process in the aganglionic hindgut of HSCR. Our real-time PCR analysis revealed inconsistencies between the mRNA levels and protein levels of genes that are associated with protein folding. This result implied that the changes for Fn and Sox10 in the aganglionic hindgut were not at the mRNA level. The discovery of noncoding RNA has revealed that RNA is not merely an intermediary between DNA and protein. mRNA is not always translated into protein (9). The inconsistency between mRNA and protein expression for Fn and Sox10 might be caused by posttranscriptional regulation. Sheehy et al. (39) have shown that the neural crest-enriched miRNA miR-452 regulates non-cell autonomous signaling involving Wnt5a, Shh, and Fgf8 that converge on Dlx2 regulation in PA1.These results suggest that posttranscriptional regulation by miRNAs is required for ENCC differentiation and ENS formation. This finding could explain the changes in Fn and Sox10 protein expression that are correlated with invariant mRNA expression in the aganglionic hindgut of HSCR and further suggests that the regulation of Fn and Sox10 was at the protein level.
A major shortcoming of this proteomic study is that only the most abundant proteins were detected, whereas the low-abundance proteins were analyzed to a limited degree. This is a technical limitation that can be addressed by the verification of as many as protein spots as possible and by technical innovation. In our future studies, we will analyze as many protein spots as possible to permit the identification of more functional proteins that are associated with HSCR.
MS-based 2DE can help to identify pathological relevant proteins in HSCR; it defines an extensive protein catalog of the normal and aganglionic hindgut and may constitute the basis to understand pathophysiological mechanisms related to the HSCR.
Methods
Sample Collection and Protein Preparation
This study was approved by the Ethics Committee of China Medical University. Written consent was obtained from the parents of the patients to use tissue that was removed during operations. The experimental groups were set as normal, ganglionic and aganglionic. The samples in the normal group were obtained from the normal colon of the patients for reasons other than HSCR (e.g., colonic trauma and colonic perforation) (n = 4), the age of these four normal controls was 11, 18, 25, and 42 mo respectively, and the mean age was 24 mo. The samples for the ganglionic and aganglionic groups were obtained from rectosigmoid HSCR patients (n = 6 for each group), and there were no patients with syndromes or long-segment HSCR in this cohort. The age of these six HSCR patients ranged from 9 to 46 mo with a mean age of 25.2 mo. None of the subjects had undergone a colostomy or a total parenteral nutrition prior to sample collection.
The full thickness of the colonic tissue was removed, immediately immersed in liquid nitrogen, and stored at −80 °C prior to protein and immunohistochemistry analyses. The frozen tissue was homogenized by FastPrep (QBIOgene, Shanghai, China) centrifugation in a lysing matrix D tube filled with 500 ml homogenization buffer containing 8 mol/l urea (Merck, Darmstadt, Germany), 2% 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS), one protease inhibitor tablet (Complete Mini; Roche, Switzerland), and dithiothreitol (3 mg/ml; Merck) for 2 × 20 s at force 4 and 1 × 20 s at force 6. The tubes were then centrifuged for 5 min at 1,000g to remove the cell debris. The supernatant was collected, and the protein concentration was determined using the protein assay kit from Bio-Rad.
2DE, Image Analysis, and Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF-MS)
All solutions and buffers were prepared following the protocols of Amersham Biosciences (Uppsala, Sweden), and the procedures were carried out as previously described (14). Briefly, 200 µg protein was first loaded onto an immobilized pH gradient strip (24 cm, pH 3–10, GE Healthcare, Uppsala, Sweden). For first-dimension isoelectric focusing, the immobilized pH gradient strip was rehydrated with 450 µl solubilized sample at 30 V for 12 h on an immobilized pH gradientphor unit (GE Healthcare). The immobilized pH gradient strips were equilibrated in 10 ml equilibration solution (6 mol/l urea, 30% glycerol, 2% sodium dodecyl sulfate (SDS), 115 mmol/l Tris–HCl pH 8.8, 20 mmol/l dithiothreitol) for 15 min and subsequently equilibrated in the same solution containing 100 mmol/l iodoacetamide rather than dithiothreitol. SDS-PAGE was performed using 12.5% polyacrylamide gels and the Ettan DALT Twelve system (GE Healthcare, Shanghai, China). Subsequently, the gels were stained with mcCBB G-250 and were scanned using a PowerLook 2100XL image scanner (Umax, Taiwan, China). Spot detection, quantification, and matching were performed using ImageMaster 2-D platinum 6.0 software (GE Healthcare) with the CBB-stained gels. The relative volume of spots (% volume calculated as the spot volume normalized as a percentage of the total volume of all the spots present in a gel) was obtained from three parallel experiments. The comparisons were intrapatient between ganglionic and aganglionic samples and were one by one between normal and aganglionic samples. The identical spots among all the HSCR samples showing at least a twofold difference were defined as differentially expressed proteins and were excised for further analysis.
Spot selection was performed manually. Digested peptides were extracted from the gel fragments and were dissolved in a MALDI matrix (5 mg/ml α-cyano-4-hydroxycinnamic acid in 0.1% trifluoroacetic acid and 50% acetonitrile), spotted onto 192-well stainless steel MALDI target plates, and analyzed using an ABI 4800 Proteomics Analyzer MALDI-TOF/TOF mass spectrometer (Applied Biosystems, Foster City, CA). The MS and MS/MS spectra were searched against the International Protein Index (IPI) human database, version 3.18, using the GPS Explorer TM v3.0 and MASCOT database search algorithms (version 2.0), with search criteria that included trypsin specificity, cysteine carbamidomethylation (C) and methionine oxidation (M) as variable modifications, one allowed trypsin missed cleavage, and a 0.2 Da MS tolerance and 0.3 Da MS/MS tolerance. Protein identifications were accepted at a Mowse score >58 and P < 0.05.
Western Blotting
The protein extract samples of 100 µg were separated by performing 12.5% SDS-PAGE and subsequently transferred in Tris–HCl methanol (20 mmol/l Tris, 150 mmol/l glycine, 20% methanol) onto polyvinylidene difluoride membranes (Millipore, Beijing, China) using a trans-blot electrophoresis transfer cell (Bio-Rad Laboratories, Richmond, CA). The blot was probed with antibodies against selected proteins or actin (Santa Cruz Biotechnology, Shanghai, China). Antigen–antibody complexes were visualized using enhanced chemiluminescence reagents. Detected bands were quantified using the Gel-pro 4.0 software (Media Cybernetics, LP), and the relative density of each protein band was calculated by dividing the optical density value of each protein by that of the loading control (actin).
Real-Time Quantitative PCR
Total RNA was extracted from the frozen tissues using Trizol reagent (Invitrogen, Shanghai, China) according to the manufacturer’s protocol. The cDNA synthesis was performed using 3 µg RNA with the TaKaRa RNA PCR kit (Takara, Dalian, China). Real-time PCR amplifications were performed in triplicate on a Light Cycler (Roche Applied Science) with the primers described in Table 3 . The housekeeping gene β-actin (Takara DR3783) served as an endogenous control.
Statistical Analysis
The data are expressed as the means ± SD of the volume (%) of spots in the 2DE analysis, relative density of bands in the western blot analysis, and 2−ΔΔCt value of each sample obtained from real-time PCR (as parametric data). Statistical significance was assessed between the normal and aganglionic group using Student’s t-test, and values of P < 0.05 were considered to be statistically significant.
Statement of Financial Support
The study was supported by the National Natural Science Foundation of China (no. 30700917) and the Science Research Project of Liaoning Key Laboratory of Congenital Malformations (no. L2013290).
Disclosure
We declare that there are no disclosures and potential conflicts of interest in this study.
References
Kenny SE, Tam PK, Garcia-Barcelo M . Hirschsprung’s disease. Semin Pediatr Surg 2010;19:194–200.
Burzynski G, Shepherd IT, Enomoto H . Genetic model system studies of the development of the enteric nervous system, gut motility and Hirschsprung’s disease. Neurogastroenterol Motil 2009;21:113–27.
Obermayr F, Hotta R, Enomoto H, Young HM . Development and developmental disorders of the enteric nervous system. Nat Rev Gastroenterol Hepatol 2013;10:43–57.
Le Douarin NM, Kalcheim C . The Neural Crest, 2nd revised edition. Cambridge: Cambridge University Press, 2009.
Goldstein AM, Hofstra RM, Burns AJ . Building a brain in the gut: development of the enteric nervous system. Clin Genet 2013;83:307–16.
Takenouchi T, Nakazawa M, Kanemura Y, et al. Hydrocephalus with Hirschsprung disease: severe end of X-linked hydrocephalus spectrum. Am J Med Genet A 2012;158A:812–5.
Vohra BP, Tsuji K, Nagashimada M, et al. Differential gene expression and functional analysis implicate novel mechanisms in enteric nervous system precursor migration and neuritogenesis. Dev Biol 2006;298:259–71.
Saeed A, Barreto L, Neogii SG, Loos A, McFarlane I, Aslam A . Identification of novel genes in Hirschsprung disease pathway using whole genome expression study. J Pediatr Surg 2012;47:303–7.
Ambros V . microRNAs: tiny regulators with great potential. Cell 2001;107:823–6.
Stroncek DF, Burns C, Martin BM, Rossi L, Marincola FM, Panelli MC . Advancing cancer biotherapy with proteomics. J Immunother 2005;28:183–92.
Swami M . Proteomics: a discovery strategy for novel cancer biomarkers. Nat Rev Cancer 2010;10:597.
Hirsch J, Hansen KC, Burlingame AL, Matthay MA . Proteomics: current techniques and potential applications to lung disease. Am J Physiol Lung Cell Mol Physiol 2004;287:L1–23.
Kikuchi T, Carbone DP . Proteomics analysis in lung cancer: challenges and opportunities. Respirology 2007;12:22–8.
Fan Y, Wang L, Zhou F, et al. Comparative proteomics of spinal cords of rat fetuses with spina bifida aperta. J Proteomics 2011;75:668–76.
Hagl CI, Thil O, Holland-Cunz S, et al. Proteome analysis of isolated myenteric plexus reveals significant changes in protein expression during postnatal development. Auton Neurosci 2005;122:1–8.
Marvin-Guy L, Lopes LV, Affolter M, et al. Proteomics of the rat gut: analysis of the myenteric plexus-longitudinal muscle preparation. Proteomics 2005;5:2561–9.
Gao H, He X, Wu M, et al. Proteomic analysis of differentially expressed proteins between stenotic and normal colon segment tissues derived from patients with Hirschsprung’s disease. Protein J 2011;30:138–42.
Bondurand N, Fouquet V, Baral V, et al. Alu-mediated deletion of SOX10 regulatory elements in Waardenburg syndrome type 4. Eur J Hum Genet 2012;20:990–4.
Chaoui A, Watanabe Y, Touraine R, et al. Identification and functional analysis of SOX10 missense mutations in different subtypes of Waardenburg syndrome. Hum Mutat 2011;32:1436–49.
Leon TY, Ngan ES, Poon HC, et al. Transcriptional regulation of RET by Nkx2-1, Phox2b, Sox10, and Pax3. J Pediatr Surg 2009;44:1904–12.
Pelisch F, Khauv D, Risso G, et al. Involvement of hnRNP A1 in the matrix metalloprotease-3-dependent regulation of Rac1 pre-mRNA splicing. J Cell Biochem 2012;113:2319–29.
Clower CV, Chatterjee D, Wang Z, Cantley LC, Vander Heiden MG, Krainer AR . The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci USA 2010;107:1894–9.
Pan H, Luo C, Li R, et al. Cyclophilin A is required for CXCR4-mediated nuclear export of heterogeneous nuclear ribonucleoprotein A2, activation and nuclear translocation of ERK1/2, and chemotactic cell migration. J Biol Chem 2008;283:623–37.
Guha M, Pan H, Fang JK, Avadhani NG . Heterogeneous nuclear ribonucleoprotein A2 is a common transcriptional coactivator in the nuclear transcription response to mitochondrial respiratory stress. Mol Biol Cell 2009;20:4107–19.
Tauler J, Zudaire E, Liu H, Shih J, Mulshine JL . hnRNP A2/B1 modulates epithelial-mesenchymal transition in lung cancer cell lines. Cancer Res 2010;70:7137–47.
Wolf K, Friedl P . Extracellular matrix determinants of proteolytic and non-proteolytic cell migration. Trends Cell Biol 2011;21:736–44.
Breau MA, Pietri T, Eder O, et al. Lack of beta1 integrins in enteric neural crest cells leads to a Hirschsprung-like phenotype. Development 2006;133:1725–34.
Parikh DH, Tam PK, Van Velzen D, Edgar D . The extracellular matrix components, tenascin and fibronectin, in Hirschsprung’s disease: an immunohistochemical study. J Pediatr Surg 1994;29:1302–6.
Miner JH, Yurchenco PD . Laminin functions in tissue morphogenesis. Annu Rev Cell Dev Biol 2004;20:255–84.
Fujimoto T, Hata J, Yokoyama S, Mitomi T . A study of the extracellular matrix protein as the migration pathway of neural crest cells in the gut: analysis in human embryos with special reference to the pathogenesis of Hirschsprung’s disease. J Pediatr Surg 1989;24:550–6.
Kadrmas JL, Beckerle MC . The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol 2004;5:920–31.
Stölting M, Wiesner C, van Vliet V, et al. Lasp-1 regulates podosome function. PLoS ONE 2012;7:e35340.
Luo G, Herrera AH, Horowits R . Molecular interactions of N-RAP, a nebulin-related protein of striated muscle myotendon junctions and intercalated disks. Biochemistry 1999;38:6135–43.
Hubbi ME, Gilkes DM, Baek JH, Semenza GL . Four-and-a-half LIM domain proteins inhibit transactivation by hypoxia-inducible factor 1. J Biol Chem 2012;287:6139–49.
Zhong Y, Zhu J, Wang Y, et al. LIM domain protein TES changes its conformational states in different cellular compartments. Mol Cell Biochem 2009;320:85–92.
Wang JX, Qin P, Liu QL, et al. Detection and significance of serum protein marker of Hirschsprung disease. Pediatrics 2007;120:e56–60.
Rozenblum GT, Gimona M . Calponins: adaptable modular regulators of the actin cytoskeleton. Int J Biochem Cell Biol 2008;40:1990–5.
Park K, Choi K, Kim H, et al. Isoflavone-deprived soy peptide suppresses mammary tumorigenesis by inducing apoptosis. Exp Mol Med 2009;41:371–81.
Sheehy NT, Cordes KR, White MP, Ivey KN, Srivastava D . The neural crest-enriched microRNA miR-452 regulates epithelial-mesenchymal signaling in the first pharyngeal arch. Development 2010;137:4307–16.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, SC., Chen, F., Jiang, KL. et al. Comparative proteomic profiles of the normal and aganglionic hindgut in human Hirschsprung disease. Pediatr Res 75, 754–761 (2014). https://doi.org/10.1038/pr.2014.33
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/pr.2014.33
This article is cited by
-
Mutations in Smad-interacting protein 1 gene are responsible for absence of its expression in Hirschsprung’s disease
Clinical and Experimental Medicine (2018)
-
Postnatal human enteric neuronal progenitors can migrate, differentiate, and proliferate in embryonic and postnatal aganglionic gut environments
Pediatric Research (2017)