Abstract
Background:
Few genetic variants have been confirmed as being associated with prostate cancer-specific mortality (PCSM). A recent study identified 22 candidate single-nucleotide polymorphisms (SNPs) associated with PCSM in a Seattle-based patient cohort. Five of these associations were replicated in an independent Swedish cohort.
Methods:
We genotyped these 22 SNPs in Physicians’ Health Study (PHS) participants diagnosed with prostate cancer (PCa). Using the same model that was found to be most significant in the Seattle cohort, we examined the association of these SNPs with lethal disease with Cox proportional hazards models.
Results:
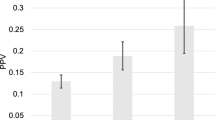
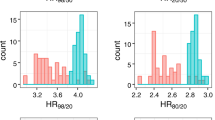
One SNP, rs5993891 in the ARVCF gene on chromosome 22q11, which had also replicated in the Swedish cohort, was also significantly associated with PCSM in the PHS cohort (hazard ratio (HR)=0.32; P=0.01). When we tested this SNP in an additional cohort (Health Professionals Follow-up Study, HPFS), the association was null (HR=0.95, P=0.90); however, a meta-analysis across all studies showed a statistically significant association with a HR of 0.52 (0.29–0.93, P=0.03).
Conclusions:
The association of rs5993891 with PCSM was further replicated in PHS and remains significant in a meta-analysis, though there was no association in HPFS. This SNP may contribute to a genetic panel of SNPs to determine at diagnosis whether a patient is more likely to exhibit an indolent or aggressive form of PCa. This study also emphasizes the importance of multiple rounds of replication.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 4 print issues and online access
$259.00 per year
only $64.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Lin DW, FitzGerald LM, Fu R, Kwon EM, Zheng SL, Kolb S et al. Genetic variants in the LEPR, CRY1, RNASEL, IL4, and ARVCF genes are prognostic markers of prostate cancer-specific mortality. Cancer Epidemiol Biomark Prev 2011; 20: 1928–1936.
Steering Committee of the Physicians’ Health Study Research Group. Final report on the aspirin component of the ongoing Physicians’ Health Study. N Engl J Med 1989; 321: 129–135.
Penney KL, Schumacher FR, Li H, Kraft P, Morris JS, Kurth T et al. A large prospective study of SEP15 genetic variation, interaction with plasma selenium levels, and prostate cancer risk and survival. Cancer Prev Res 2010; 3: 604–610.
Shui IM, Mucci LA, Kraft P, Tamimi RM, Lindstrom S, Penney KL et al. Vitamin D-related genetic variation, plasma vitamin D and risk of lethal prostate cancer: a prospective nested case-control study. J Natl Cancer Inst 2012; 104: 690–699.
Roberts DG, Morrison TB, Liu-Cordero SN, Cho J, Garcia J, Kanigan TS et al. A nanoliter fluidic platform for large-scale single nucleotide polymorphism genotyping. Biotechniques 2009; 46 (Suppl 3): ix–xiii.
Mariner DJ, Wang J, Reynolds AB . ARVCF localizes to the nucleus and adherens junctions and is mutually exclusive with p120(ctn) in E-cadherin complexes. J Cell Sci 2000; 113 (Pt. 8): 1481–1490.
Reynolds AB, Roczniak-Fergusion A . Emerging roles for p120-catenin in cell adhesion and cancer. Oncogene 2004; 23: 7947–7956.
Reintsch WE, Mandato CA, McCrea PD, Fagotto F . Inhibition of cell adhesion by xARVCF indicate a regulatory function at the plasma membrane. Dev Dyn 2008; 237: 2328–2341.
Acknowledgements
We are grateful to the participants and staff of the Physicians’ Health Study and Health Professionals Follow-up Study for their valuable contributions, as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA and WY. In addition, this study was approved by the Connecticut Department of Public Health (DPH) Human Investigations Committee. Certain data used in this publication were obtained from the DPH. This work was supported by the Prostate Cancer Foundation. The Physicians’ Health Study was supported by grants CA34944, CA40360 and CA097193 from the National Cancer Institute and HL26490 and HL34595 from the National Heart, Lung and Blood Institute. The Health Professionals Follow-up Study was supported by grants R01CA133891, R01CA141298-02 and P01CA055075. KLP is supported by a Prostate Cancer Foundation Young Investigator Award. IMS is supported by a US Army Department of Defense Prostate Cancer Research Program Post-doctoral Fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Prostate Cancer and Prostatic Diseases website
Supplementary information
Rights and permissions
About this article
Cite this article
Penney, K., Shui, I., Feng, Z. et al. Replication of a genetic variant for prostate cancer-specific mortality. Prostate Cancer Prostatic Dis 18, 260–263 (2015). https://doi.org/10.1038/pcan.2015.18
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/pcan.2015.18
This article is cited by
-
Germline variants in IL4, MGMT and AKT1 are associated with prostate cancer-specific mortality: An analysis of 12,082 prostate cancer cases
Prostate Cancer and Prostatic Diseases (2018)