Abstract
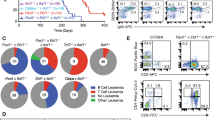
Signal transducer and activator of transcription 5 (STAT5) activation occurs frequently in human progenitor B-cell acute lymphoblastic leukemia (B-ALL). To identify gene alterations that cooperate with STAT5 activation to initiate leukemia, we crossed mice expressing a constitutively active form of STAT5 (Stat5b-CA) with mice in which a mutagenic Sleeping Beauty transposon (T2/Onc) was mobilized only in B cells. Stat5b-CA mice typically do not develop B-ALL (<2% penetrance); in contrast, 89% of Stat5b-CA mice in which the T2/Onc transposon had been mobilized died of B-ALL by 3 months of age. High-throughput sequencing approaches were used to identify genes frequently targeted by the T2/Onc transposon; these included Sos1 (74%), Kdm2a (35%), Jak1 (26%), Bmi1 (19%), Prdm14 or Ncoa2 (13%), Cdkn2a (10%), Ikzf1 (8%), Caap1 (6%) and Klf3 (6%). Collectively, these mutations target three major cellular processes: (i) the Janus kinase/STAT5 pathway (ii) progenitor B-cell differentiation and (iii) the CDKN2A tumor-suppressor pathway. Transposon insertions typically resulted in altered expression of these genes, as well as downstream pathways including STAT5, extracellular signal-regulated kinase (Erk) and p38. Importantly, expression of Sos1 and Kdm2a, and activation of p38, correlated with survival, further underscoring the role these genes and associated pathways have in B-ALL.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Inaba H, Greaves M, Mullighan CG . Acute lymphoblastic leukaemia. Lancet 2013; 381: 1943–1955.
Roberts KG, Mullighan CG . Genomics in acute lymphoblastic leukaemia: insights and treatment implications. Nat Rev Clin Oncol 2015; 12: 344–357.
Mullighan CG . Genomic characterization of childhood acute lymphoblastic leukemia. Semin Hematol 2013; 50: 314–324.
Heltemes-Harris LM, Willette MJ, Ramsey LB, Qiu YH, Neeley ES, Zhang N et al. Ebf1 or Pax5 haploinsufficiency synergizes with STAT5 activation to initiate acute lymphoblastic leukemia. J Exp Med 2011; 208: 1135–1149.
Nakayama J, Yamamoto M, Hayashi K, Satoh H, Bundo K, Kubo M et al. BLNK suppresses pre-B-cell leukemogenesis through inhibition of JAK3. Blood 2009; 113: 1483–1492.
Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA . Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature 2005; 436: 221–226.
Dupuy AJ, Rogers LM, Kim J, Nannapaneni K, Starr TK, Liu P et al. A modified Sleeping Beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Res 2009; 69: 8150–8156.
Abbott KL, Nyre ET, Abrahante J, Ho YY, Isaksson Vogel R, Starr TK . The candidate cancer gene database: a database of cancer driver genes from forward genetic screens in mice. Nucleic acids Res 2015; 43: D844–D848.
Berquam-Vrieze KE, Nannapaneni K, Brett BT, Holmfeldt L, Ma J, Zagorodna O et al. Cell of origin strongly influences genetic selection in a mouse model of T-ALL. Blood 2011; 118: 4646–4656.
Starr TK, Allaei R, Silverstein KA, Staggs RA, Sarver AL, Bergemann TL et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science 2009; 323: 1747–1750.
Hobeika E, Thiemann S, Storch B, Jumaa H, Nielsen PJ, Pelanda R et al. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci USA 2006; 103: 13789–13794.
Brett BT, Berquam-Vrieze KE, Nannapaneni K, Huang J, Scheetz TE, Dupuy AJ . Novel molecular and computational methods improve the accuracy of insertion site analysis in Sleeping Beauty-induced tumors. PloS One 2011; 6: e24668.
Sarver AL, Erdman J, Starr T, Largaespada DA, Silverstein KA . TAPDANCE: an automated tool to identify and annotate transposon insertion CISs and associations between CISs from next generation sequence data. BMC Bioinform 2012; 13: 154.
Schwickert TA, Tagoh H, Gultekin S, Dakic A, Axelsson E, Minnich M et al. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat Immunol 2014; 15: 283–293.
Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med 2009; 360: 470–480.
Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 2007; 446: 758–764.
Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature 2008; 453: 110–114.
Cheng Z, Cheung P, Kuo AJ, Yukl ET, Wilmot CM, Gozani O et al. A molecular threading mechanism underlies Jumonji lysine demethylase KDM2A regulation of methylated H3K36. Genes Dev 2014; 28: 1758–1771.
Lu T, Jackson MW, Wang B, Yang M, Chance MR, Miyagi M et al. Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci USA 2010; 107: 46–51.
Lu T, Yang M, Huang DB, Wei H, Ozer GH, Ghosh G et al. Role of lysine methylation of NF-kappaB in differential gene regulation. Proc Natl Acad Sci USA 2013; 110: 13510–13515.
Mullighan CG, Williams RT, Downing JR, Sherr CJ . Failure of CDKN2A/B (INK4A/B-ARF)-mediated tumor suppression and resistance to targeted therapy in acute lymphoblastic leukemia induced by BCR-ABL. Genes Dev 2008; 22: 1411–1415.
Williams RT, Sherr CJ . The INK4-ARF (CDKN2A/B) locus in hematopoiesis and BCR-ABL-induced leukemias. Cold Spring Harb Symp Quant Biol 2008; 73: 461–467.
Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M . The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 1999; 397: 164–168.
Jun JE, Yang M, Chen H, Chakraborty AK, Roose JP . Activation of extracellular signal-regulated kinase but not of p38 mitogen-activated protein kinase pathways in lymphocytes requires allosteric activation of SOS. Mol Cell Biol 2013; 33: 2470–2484.
Mandal M, Powers SE, Ochiai K, Georgopoulos K, Kee BL, Singh H et al. Ras orchestrates exit from the cell cycle and light-chain recombination during early B cell development. Nat Immunol 2009; 10: 1110–1117.
Diehl NL, Enslen H, Fortner KA, Merritt C, Stetson N, Charland C et al. Activation of the p38 mitogen-activated protein kinase pathway arrests cell cycle progression and differentiation of immature thymocytes in vivo. J Exp Med 2000; 191: 321–334.
Li Q, Bohin N, Wen T, Ng V, Magee J, Chen SC et al. Oncogenic Nras has bimodal effects on stem cells that sustainably increase competitiveness. Nature 2013; 504: 143–147.
Zhang J, Mullighan CG, Harvey RC, Wu G, Chen X, Edmonson M et al. Key pathways are frequently mutated in high-risk childhood acute lymphoblastic leukemia: a report from the Children's Oncology Group. Blood 2011; 118: 3080–3087.
Irving J, Matheson E, Minto L, Blair H, Case M, Halsey C et al. Ras pathway mutations are prevalent in relapsed childhood acute lymphoblastic leukemia and confer sensitivity to MEK inhibition. Blood 2014; 124: 3420–3430.
Shojaee S, Caeser R, Buchner M, Park E, Swaminathan S, Hurtz C et al. Erk negative feedback control enables pre-B cell transformation and represents a therapeutic target in acute lymphoblastic leukemia. Cancer Cell 2015; 28: 1–15.
Evelyn CR, Duan X, Biesiada J, Seibel WL, Meller J, Zheng Y . Rational design of small molecule inhibitors targeting the Ras GEF, SOS1. Chem Biol 2014; 21: 1618–1628.
Bulavin DV, Kovalsky O, Hollander MC, Fornace AJ Jr . Loss of oncogenic H-ras-induced cell cycle arrest and p38 mitogen-activated protein kinase activation by disruption of Gadd45a. Mol Cell Biol 2003; 23: 3859–3871.
Iwasa H, Han J, Ishikawa F . Mitogen-activated protein kinase p38 defines the common senescence-signalling pathway. Genes Cells 2003; 8: 131–144.
Wang W, Chen JX, Liao R, Deng Q, Zhou JJ, Huang S et al. Sequential activation of the MEK-extracellular signal-regulated kinase and MKK3/6-p38 mitogen-activated protein kinase pathways mediates oncogenic ras-induced premature senescence. Mol Cell Biol 2002; 22: 3389–3403.
Chen Z, Shojaee S, Buchner M, Geng H, Lee JW, Klemm L et al. Signalling thresholds and negative B-cell selection in acute lymphoblastic leukaemia. Nature 2015; 521: 357–361.
Cornall RJ, Cyster JG, Hibbs ML, Dunn AR, Otipoby KL, Clark EA et al. Polygenic autoimmune traits: Lyn, CD22, and SHP-1 are limiting elements of a biochemical pathway regulating BCR signaling and selection. Immunity 1998; 8: 497–508.
Vougioukalaki M, Kanellis DC, Gkouskou K, Eliopoulos AG . Tpl2 kinase signal transduction in inflammation and cancer. Cancer Lett 2011; 304: 80–89.
Vu TT, Gatto D, Turner V, Funnell AP, Mak KS, Norton LJ et al. Impaired B cell development in the absence of Kruppel-like factor 3. J Immunol 2011; 187: 5032–5042.
Lessard J, Sauvageau G . Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature 2003; 423: 255–260.
Park I-k, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 2003; 423: 302–305.
Xu H, Yang W, Perez-Andreu V, Devidas M, Fan Y, Cheng C et al. Novel susceptibility variants at 10p12.31-12.2 for childhood acute lymphoblastic leukemia in ethnically diverse populations. J Natl Cancer Inst 2013; 105: 733–742.
Oguro H, Yuan J, Ichikawa H, Ikawa T, Yamazaki S, Kawamoto H et al. Poised lineage specification in multipotential hematopoietic stem and progenitor cells by the polycomb protein Bmi1. Cell Stem Cell 2010; 6: 279–286.
Arranz L, Herrera-Merchan A, Ligos JM, de Molina A, Dominguez O, Gonzalez S . Bmi1 is critical to prevent Ikaros-mediated lymphoid priming in hematopoietic stem cells. Cell Cycle 2012; 11: 65–78.
Burchill MA, Goetz CA, Prlic M, O'Neil JJ, Harmon IR, Bensinger SJ et al. Distinct effects of STAT5 activation on CD4+ and CD8+ T cell homeostasis: development of CD4+CD25+ regulatory T cells versus CD8+ memory T cells. J Immunol 2003; 171: 5853–5864.
Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA . Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature 2005; 436: 272–276.
Starr TK, Largaespada DA . Cancer gene discovery using the Sleeping Beauty transposon. Cell Cycle 2005; 4: 1744–1748.
Acknowledgements
We thank Peter Schoettler, Chris Reis, Alyssa Kne, Amy Mack and Emilea Sykes for assistance with animal husbandry, Maya Raghunandan for technical assistance, Paul Champoux for assistance with Flow cytometry and Dr Richard Williams for helpful comments. This work was supported by a Brainstorm grant from the University of Minnesota Masonic Cancer Center (MAF and DAL), NIH grants R01 CA154998 and CA151845 to MAF, NIH grant 5R00CA151672-04 to TKS, NIH R01 CA113636 to DAL, an NIH Institutional Shared Resource grant to the Masonic Cancer Center P30-CA77598 and a Leukemia and Lymphoma Society Scholar award (MAF).
Author contributions
LMH-H, DAL and MAF designed research; LMH-H, JDL, TKS and GKH performed experiments and analyzed data; ALS and MAF analyzed data, LMH-H and MAF wrote the paper; and all authors critically reviewed and edited the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
DAL has ownership interest (including patents) in Discovery Genomics, Inc., and NeoClone Biotechnologies International and is also a consultant/advisory board member of Discovery Genomics, Inc., and NeoClone Biotechnologies International.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Heltemes-Harris, L., Larson, J., Starr, T. et al. Sleeping Beauty transposon screen identifies signaling modules that cooperate with STAT5 activation to induce B-cell acute lymphoblastic leukemia. Oncogene 35, 3454–3464 (2016). https://doi.org/10.1038/onc.2015.405
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.405
This article is cited by
-
Identification of mutations that cooperate with defects in B cell transcription factors to initiate leukemia
Oncogene (2021)
-
Signalling input from divergent pathways subverts B cell transformation
Nature (2020)
-
A Cell Density-Dependent Reporter in the Drosophila S2 Cells
Scientific Reports (2019)
-
It is a differentiation game: STAT5 in a new role
Cell Death & Differentiation (2017)
-
Antagonism of B cell enhancer networks by STAT5 drives leukemia and poor patient survival
Nature Immunology (2017)