Abstract
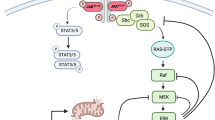
To understand the role of cytokine and growth factor receptor-mediated signaling in leukemia pathogenesis, we designed a functional RNA interference (RNAi) screen targeting 188 cytokine and growth factor receptors that we found highly expressed in primary leukemia specimens. Using this screen, we identified interleukin-2 gamma receptor (IL2Rγ) as a critical growth determinant for a JAK3A572V mutation-positive acute myeloid leukemia cell line. We observed that knockdown of IL2Rγ abrogates phosphorylation of JAK3 and downstream signaling molecules, JAK1, STAT5, MAPK and pS6 ribosomal protein. Overexpression of IL2Rγ in murine cells increased the transforming potential of activating JAK3 mutations, whereas absence of IL2Rγ completely abrogated the clonogenic potential of JAK3A572V, as well as the transforming potential of additional JAK3-activating mutations such as JAK3M511I. In addition, mutation at the IL2Rγ interaction site in the FERM domain of JAK3 (Y100C) completely abrogated JAK3-mediated leukemic transformation. Mechanistically, we found IL2Rγ contributes to constitutive JAK3 mutant signaling by increasing JAK3 expression and phosphorylation. Conversely, we found that mutant, but not wild-type JAK3, increased the expression of IL2Rγ, indicating IL2Rγ and JAK3 contribute to constitutive JAK/STAT signaling through their reciprocal regulation. Overall, we demonstrate a novel role for IL2Rγ in potentiating oncogenesis in the setting of JAK3-mutation-positive leukemia. In addition, our study highlights an RNAi-based functional assay that can be used to facilitate the identification of non-kinase cytokine and growth factor receptor targets for inhibiting leukemic cell growth.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Estey E, Dohner H Acute myeloid leukaemia. Lancet 2006; 368: 1894–1907.
Graf M, Hecht K, Reif S, Pelka-Fleischer R, Pfister K, Schmetzer H Expression and prognostic value of hemopoietic cytokine receptors in acute myeloid leukemia (AML): implications for future therapeutical strategies. Eur J Haematol 2004; 72: 89–106.
Van Etten RA Aberrant cytokine signaling in leukemia. Oncogene 2007; 26: 6738–6749.
Vainchenker W, Constantinescu SN . JAK/STAT signaling in hematological malignancies. Oncogene 2013; 32: 2601–2613.
Tefferi A . Novel mutations and their functional and clinical relevance in myeloproliferative neoplasms: JAK2, MPL, TET2, ASXL1, CBL, IDH and IKZF1. Leukemia 2010; 24: 1128–1138.
Russell LJ, Capasso M, Vater I, Akasaka T, Bernard OA, Calasanz MJ et al. Deregulated expression of cytokine receptor gene, CRLF2, is involved in lymphoid transformation in B-cell precursor acute lymphoblastic leukemia. Blood 2009; 114: 2688–2698.
Pikman Y, Lee BH, Mercher T, McDowell E, Ebert BL, Gozo M et al. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med 2006; 3: e270.
Jin L, Lee EM, Ramshaw HS, Busfield SJ, Peoppl AG, Wilkinson L et al. Monoclonal antibody-mediated targeting of CD123, IL-3 receptor alpha chain, eliminates human acute myeloid leukemic stem cells. Cell Stem Cell 2009; 5: 31–42.
O’Shea JJ, Park H, Pesu M, Borie D, Changelian P New strategies for immunosuppression: interfering with cytokines by targeting the Jak/Stat pathway. Curr Opin Reumatol 2005; 17: 305–311.
Ferretti E, Cocco C, Airoldi I, Pistoia V Targeting acute myeloid leukemia cells with cytokines. J Leukoc Biol 2012; 92: 567–575.
Majeti R, Chao MP, Alizadeh AA, Pang WW, Jaiswal S, Gibbs Jr KD et al. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell 2009; 138: 286–299.
O'Shea JJ, Pesu M, Borie DC, Changelian PS A new modality for immunosuppression: targeting the JAK/STAT pathway. Nat Rev Drug Discov 2004; 3: 555–564.
Tang W, Huo H, Zhu J, Ji H, Zou W, Xu L et al. Critical sites for the interaction between IL-2Rgamma and JAK3 and the following signaling. Biochem Biophys Res Commun 2001; 283: 598–605.
Cacalano NA, Migone TS, Bazan F, Hanson EP, Chen M, Candotti F et al. Autosomal SCID caused by a point mutation in the N-terminus of Jak3: mapping of the Jak3-receptor interaction domain. EMBO J 1999; 18: 1549–1558.
Baird AM, Thomis DC, Berg LJ T cell development and activation in Jak3-deficient mice. J Leukoc Biol 1998; 63: 669–677.
Candotti F, Oakes SA, Johnston JA, Notarangelo LD, O'Shea JJ, Blaese RM In vitro correction of JAK3-deficient severe combined immunodeficiency by retroviral-mediated gene transduction. J Exp Med 1996; 183: 2687–2692.
Bellanger D, Jacquemin V, Chopin M, Pierron G, Bernard OA, Ghysdael J et al. Recurrent JAK1 and JAK3 somatic mutations in T-cell prolymphocytic leukemia. Leukemia 2014; 28: 417–419.
De Vita S, Mulligan C, McElwaine S, Dagna-Bricarelli F, Spinelli M, Basso G et al. Loss-of-function JAK3 mutations in TMD and AMKL of Down syndrome. Br J Haematol 2007; 137: 337–341.
Jeong EG, Kim MS, Nam HK, Min CK, Lee S, Chung YJ et al. Somatic mutations of JAK1 and JAK3 in acute leukemias and solid cancers. Clin Cancer Res 2008; 14: 3716–3721.
Kiyoi H, Yamaji S, Kojima S, Naoe T JAK3 mutations occur in acute megakaryoblastic leukemia both in Down syndrome children and non-Down syndrome adults. Leukemia 2007; 21: 574–576.
Sakaguchi H, Okuno Y, Muramatsu H, Yoshida K, Shiraishi Y, Takahashi M et al. Exome sequencing identifies secondary mutations of SETBP1 and JAK3 in juvenile myelomonocytic leukemia. Nat Genet 2013; 45: 937–941.
Sato T, Toki T, Kanezaki R, Xu G, Terui K, Kanegane H et al. Functional analysis of JAK3 mutations in transient myeloproliferative disorder and acute megakaryoblastic leukaemia accompanying Down syndrome. Br J Haematol 2008; 141: 681–688.
Walters DK, Mercher T, Gu TL, O’Hare T, Tyner JW, Loriaux M et al. Activating alleles of JAK3 in acute megakaryoblastic leukemia. Cancer Cell 2006; 10: 65–75.
Bains T, Heinrich MC, Loriaux MM, Beadling C, Nelson D, Warrick A et al. Newly described activating JAK3 mutations in T-cell acute lymphoblastic leukemia. Leukemia 2012; 26: 2144–2146.
Blink M, Buitenkamp TD, van den Heuvel-Eibrink MM, Danen-van Oorschot AA, de Haas V, Reinhardt D et al. Frequency and prognostic implications of JAK 1-3 aberrations in Down syndrome acute lymphoblastic and myeloid leukemia. Leukemia 2011; 25: 1365–1368.
Tefferi A . JAK and MPL mutations in myeloid malignancies. Leuk Lymphoma 2008; 49: 388–397.
Constantinescu SN, Girardot M, Pecquet C Mining for JAK-STAT mutations in cancer. Trends Biochem Sci 2008; 33: 122–131.
Murphy JM, Young IG . IL-3, IL-5, and GM-CSF signaling: crystal structure of the human beta-common receptor. Vitam Horm 2006; 74: 1–30.
Hercus TR, Dhagat U, Kan WL, Broughton SE, Nero TL, Perugini M et al. Signalling by the betac family of cytokines. Cytokine Growth Factor Rev 2013; 24: 189–201.
Funakoshi-Tago M, Pelletier S, Moritake H, Parganas E, Ihle JN Jak2 FERM domain interaction with the erythropoietin receptor regulates Jak2 kinase activity. Mol Cell Biol 2008; 28: 1792–1801.
Spivak JL, Merchant A, Williams DM, Ophelia Rogers O, Zhao W, Moliterno AR et al. A Functional Thrombopoietin Receptor Is Required for Full Expression of Phenotype in a JAK2 V617F Transgenic Mouse Model of Polycythemia Vera. Am Soc Hematol Blood 2012; 120: 427.
Hofmann SR, Lam AQ, Frank S, Zhou YJ, Ramos HL, Kanno Y et al. Jak3-independent trafficking of the common gamma chain receptor subunit: chaperone function of Jaks revisited. Mol Cell Biol 2004; 24: 5039–5049.
Cornejo MG, Boggon TJ, Mercher T JAK3: a two-faced player in hematological disorders. Int J Biochem Cell Biol 2009; 41: 2376–2379.
Ju W, Zhang M, Jiang JK, Thomas CJ, Oh U, Bryant BR et al. CP-690,550, a therapeutic agent, inhibits cytokine-mediated Jak3 activation and proliferation of T cells from patients with ATL and HAM/TSP. Blood 2011; 117: 1938–1946.
Vijayakrishnan L, Venkataramanan R, Gulati P Treating inflammation with the Janus kinase inhibitor CP-690,550. Trends Pharmacol Sci 2011; 32: 25–34.
Kontzias A, Kotlyar A, Laurence A, Changelian P, O'Shea JJ Jakinibs: a new class of kinase inhibitors in cancer and autoimmune disease. Curr Opin Pharmacol 2012; 12: 464–470.
Wernig G, Gonneville JR, Crowley BJ, Rodrigues MS, Reddy MM, Hudon HE et al. The Jak2V617F oncogene associated with myeloproliferative diseases requires a functional FERM domain for transformation and for expression of the Myc and Pim proto-oncogenes. Blood 2008; 111: 3751–3759.
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003; 4: 249–264.
Rubin JB Chemokine signaling in cancer: one hump or two? Semin Cancer Biol 2009; 19: 116–122.
Kiss C, Benko I, Kovacs P Leukemic cells and the cytokine patchwork. Pediatr Blood Cancer 2004; 42: 113–121.
Tyner JW, Walters DK, Willis SG, Luttropp M, Oost J, Loriaux M et al. RNAi screening of the tyrosine kinome identifies therapeutic targets in acute myeloid leukemia. Blood 2008; 111: 2238–2245.
Acknowledgements
We thank Sarah Bowden for administrative support. AA is supported by a National Cancer Institute Career Development Award (5 K99 CA151670-02), Collins Foundation, Knight Pilot Project and Friends of Doernbecher grants. JWT is supported by grants from the V-Foundation, Gabrielle’s Angel Foundation and the National Cancer Institute (5R00CA151457-04; 1R01CA183947-01). BJD is a Howard Hughes Medical Institute investigator. This work is also supported by The Leukemia & Lymphoma Society. We thank Dr. Jamie Keck (Dr. Brian Druker’s Laboratory, OHSU) for providing her technical assistance in creating Gateway compatible pMXs-IRES-Puromycin vector.
Author Contributions
AA, RJM, KW-S, RMB and MAD designed the research, performed experiments and wrote the paper. SM-T and BP helped with microarray gene expression analysis. JWT, CET, CAE and BJD provided critical feedback and helped with manuscript preparation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
OHSU and BJD have a financial interest in Molecular MD (5% equity or less). This potential conflict of interest has been reviewed and managed by the OHSU Conflict of Interest in Research Committee and the Integrity Program Oversight Council. OHSU has clinical trial contracts with Novartis, and Bristol Myers Squibb to pay for patient costs, nurse and data manager salaries and institutional overhead. BJD does not derive salary, nor do their laboratories receive funds, from these contracts. BJD serves as a consultant for Roche and Nodality (Consulting income $10,000 or over).
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Agarwal, A., MacKenzie, R., Eide, C. et al. Functional RNAi screen targeting cytokine and growth factor receptors reveals oncorequisite role for interleukin-2 gamma receptor in JAK3-mutation-positive leukemia. Oncogene 34, 2991–2999 (2015). https://doi.org/10.1038/onc.2014.243
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2014.243
This article is cited by
-
In vivo impact of JAK3 A573V mutation revealed using zebrafish
Cellular and Molecular Life Sciences (2022)
-
Integrated genomic analysis identifies clinically relevant subtypes of renal clear cell carcinoma
BMC Cancer (2018)