Abstract
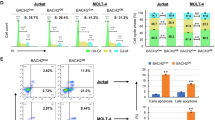
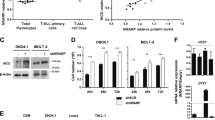
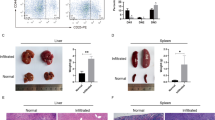
Notch signaling is a highly conserved cell–cell communication pathway regulating normal development and tissue homeostasis. Aberrant Notch signaling represents an important oncogenic mechanism for T cell acute lymphoblastic leukemia (T-ALL), an aggressive subset of the most common malignant childhood cancer ALL. Therefore, understanding the molecular regulation of Notch signaling is critical to identify new approaches to block aberrant Notch oncogenic activity. The family of three MAML transcriptional coactivators is crucial for Notch signaling activation. The prototypic member MAML1 is the major coactivator that regulates Notch oncogenic activities in leukemic cells. However, the molecular basis underlying MAML1 coactivator function that contributes to Notch signaling remains unclear. In this study, we performed proteomic studies and identified DDX5, an ATP-dependent DEAD-box RNA helicase, as a component of the MAML1 protein complex. DDX5 interacts with MAML1 in vitro and in vivo, and is associated with the endogenous NOTCH1 transcription activation complex in human T-ALL leukemic cells. Lentivirus-mediated short-hairpin RNA knock-down of DDX5 resulted in decreased expression of Notch target genes, reduced cell proliferation and increased apoptosis in cultured human leukemic cells with constitutive activation of Notch signaling. Also, DDX5 depletion inhibited the growth of human leukemia xenograft in nude mice. Moreover, DDX5 is highly expressed in primary human T-ALL leukemic cells based on the analyses of Oncomine and GEO databases, and Immunohistochemical staining. Our overall findings revealed a critical role of DDX5 in promoting efficient Notch-mediated transcription in leukemic cells, suggesting that DDX5 might be critical for NOTCH1-mediated T-ALL pathogenesis and thus is a potential new target for modulating the Notch signaling in leukemia.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Artavanis-Tsakonas S, Muskavitch MA . Notch: the past, the present, and the future. Curr Top Dev Biol 2010; 92: 1–29.
Koch U, Radtke F . Notch signaling in solid tumors. Curr Top Dev Biol 2010; 92: 411–455.
Paganin M, Ferrando A . Molecular pathogenesis and targeted therapies for NOTCH1-induced T-cell acute lymphoblastic leukemia. Blood Rev 2011; 25: 83–90.
Aster JC, Blacklow SC, Pear WS . Notch signalling in T-cell lymphoblastic leukaemia/lymphoma and other haematological malignancies. J Pathol 2011; 223: 262–273.
Weng AP, Ferrando AA, Lee W, JPt Morris, Silverman LB, Sanchez-Irizarry C et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004; 306: 269–271.
Chiang MY, Xu L, Shestova O, Histen G, L'Heureux S, Romany C et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest 2008; 118: 3181–3194.
Ranganathan P, Weaver KL, Capobianco AJ . Notch signalling in solid tumours: a little bit of everything but not all the time. Nat Rev Cancer 2011; 11: 338–351.
Kopan R, Ilagan MX . The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 2009; 137: 216–233.
Wu L, Aster JC, Blacklow SC, Lake R, Artavanis-Tsakonas S, Griffin JD . MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat Genet 2000; 26: 484–489.
Lin SE, Oyama T, Nagase T, Harigaya K, Kitagawa M . Identification of new human mastermind proteins defines a family that consists of positive regulators for notch signaling. J Biol Chem 2002; 277: 50612–50620.
Wu L, Sun T, Kobayashi K, Gao P, Griffin JD . Identification of a family of mastermind-like transcriptional coactivators for mammalian notch receptors. Mol Cell Biol 2002; 22: 7688–7700.
McElhinny AS, Li JL, Wu L . Mastermind-like transcriptional co-activators: emerging roles in regulating cross talk among multiple signaling pathways. Oncogene 2008; 27: 5138–5147.
Ford MJ, Anton IA, Lane DP . Nuclear protein with sequence homology to translation initiation factor eIF-4A. Nature 1988; 332: 736–738.
Hirling H, Scheffner M, Restle T, Stahl H . RNA helicase activity associated with the human p68 protein. Nature 1989; 339: 562–564.
Fuller-Pace FV, Ali S . The DEAD box RNA helicases p68 (Ddx5) and p72 (Ddx17): novel transcriptional co-regulators. Biochem Soc Trans 2008; 36 (Part 4): 609–612.
Fuller-Pace FV, Moore HC . RNA helicases p68 and p72: multifunctional proteins with important implications for cancer development. Future Oncol 2011; 7: 239–251.
Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC et al. Direct inhibition of the NOTCH transcription factor complex. Nature 2009; 462: 182–188.
Fryer CJ, White JB, Jones KA . Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol Cell 2004; 16: 509–520.
Fryer CJ, Lamar E, Turbachova I, Kintner C, Jones KA . Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev 2002; 16: 1397–1411.
Hansson ML, Popko-Scibor AE, Saint Just Ribeiro M, Dancy BM, Lindberg MJ, Cole PA et al. The transcriptional coactivator MAML1 regulates p300 autoacetylation and HAT activity. Nucleic Acids Res 2009; 37: 2996–3006.
Saint Just Ribeiro M, Hansson ML, Lindberg MJ, Popko-Scibor AE, Wallberg AE . GSK3beta is a negative regulator of the transcriptional coactivator MAML1. Nucleic Acids Res 2009; 37: 6691–7600.
Haferlach T, Kohlmann A, Wieczorek L, Basso G, Kronnie GT, Bene MC et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group. J Clin Oncol 2010; 28: 2529–2537.
Homminga I, Pieters R, Langerak AW, de Rooi JJ, Stubbs A, Verstegen M et al. Integrated transcript and genome analyses reveal NKX2-1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell 2011; 19: 484–497.
Van Vlierberghe P, Ambesi-Impiombato A, Perez-Garcia A, Haydu JE, Rigo I, Hadler M et al. ETV6 mutations in early immature human T cell leukemias. J Exp Med 2011; 208: 2571–2579.
Caretti G, Lei EP, Sartorelli V . The DEAD-box p68/p72 proteins and the noncoding RNA steroid receptor activator SRA: eclectic regulators of disparate biological functions. Cell Cycle 2007; 6: 1172–1176.
Nicol SM, Fuller-Pace FV . Analysis of the RNA helicase p68 (Ddx5) as a transcriptional regulator. Methods Mol Biol 2010; 587: 265–279.
Clark EL, Coulson A, Dalgliesh C, Rajan P, Nicol SM, Fleming S et al. The RNA helicase p68 is a novel androgen receptor coactivator involved in splicing and is overexpressed in prostate cancer. Cancer Res 2008; 68: 7938–7946.
Endoh H, Maruyama K, Masuhiro Y, Kobayashi Y, Goto M, Tai H et al. Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor alpha. Mol Cell Biol 1999; 19: 5363–5372.
Caretti G, Schiltz RL, Dilworth FJ, Di Padova M, Zhao P, Ogryzko V et al. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev Cell 2006; 11: 547–560.
Jensen ED, Niu L, Caretti G, Nicol SM, Teplyuk N, Stein GS et al. p68 (Ddx5) interacts with Runx2 and regulates osteoblast differentiation. J Cell Biochem 2008; 103: 1438–1451.
Germann S, Gratadou L, Zonta E, Dardenne E, Gaudineau B, Fougère M et al. Dual role of the ddx5/ddx17 RNA helicases in the control of the pro-migratory NFAT5 transcription factor. Oncogene 2012; 31: 4536–4549.
Janknecht R . Multi-talented DEAD-box proteins and potential tumor promoters: p68 RNA helicase (DDX5) and its paralog, p72 RNA helicase (DDX17). Am J Transl Res 2010; 2: 223–234.
Yang L, Lin C, Sun SY, Zhao S, Liu ZR . A double tyrosine phosphorylation of P68 RNA helicase confers resistance to TRAIL-induced apoptosis. Oncogene 2007; 26: 6082–6092.
Cohen AA, Geva-Zatorsky N, Eden E, Frenkel-Morgenstern M, Issaeva I, Sigal A et al. Dynamic proteomics of individual cancer cells in response to a drug. Science 2008; 322: 1511–1516.
Guo D, Ye J, Dai J, Li L, Chen F, Ma D et al. Notch-1 regulates Akt signaling pathway and the expression of cell cycle regulatory proteins cyclin D1, CDK2 and p21 in T-ALL cell lines. Leuk Res 2009; 33: 678–685.
Weng AP, Nam Y, Wolfe MS, Pear WS, Griffin JD, Blacklow SC et al. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol Cell Biol 2003; 23: 655–664.
Gu Y, Lin S, Li JL, Nakagawa H, Chen Z, Jin B et al. Altered LKB1/CREB-regulated transcription co-activator (CRTC) signaling axis promotes esophageal cancer cell migration and invasion. Oncogene 2012; 31: 469–479.
Acknowledgements
We thank Dr Vittorio Sartorelli for kindly providing us with DDX5 expression constructs, Marda Jorgensen for helping with immunohistochemical staining and Jameson Kuang for careful reading of the manuscript. This work was supported by the University of Florida Shands Cancer Center Startup fund, STOP! Children’s Cancer, Inc., and Florida Bankhead Coley Cancer Research Program (2BB11 and 1BF04 to LW). HS was supported by the National Natural Science Foundation of China (81170891). MJY was supported by NIH/NCI RO1 CA164346, Developmental Research Awards in Leukemia SPORE CA100632, UT MDACC Physician Scientist Award, American Cancer Society IRG, UT MDACC IRG and Ladies Leukemia League.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Lin, S., Tian, L., Shen, H. et al. DDX5 is a positive regulator of oncogenic NOTCH1 signaling in T cell acute lymphoblastic leukemia. Oncogene 32, 4845–4853 (2013). https://doi.org/10.1038/onc.2012.482
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2012.482
Keywords
This article is cited by
-
Targeting Notch and EGFR signaling in human mucoepidermoid carcinoma
Signal Transduction and Targeted Therapy (2021)
-
The DEAD-box protein family of RNA helicases: sentinels for a myriad of cellular functions with emerging roles in tumorigenesis
International Journal of Clinical Oncology (2021)
-
DDX5-targeting fully human monoclonal autoantibody inhibits proliferation and promotes differentiation of acute promyelocytic leukemia cells by increasing ROS production
Cell Death & Disease (2020)
-
Super enhancer inhibitors suppress MYC driven transcriptional amplification and tumor progression in osteosarcoma
Bone Research (2018)
-
DDX59 promotes DNA replication in lung adenocarcinoma
Cell Death Discovery (2017)