Abstract
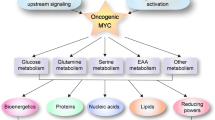
Cell proliferation requires the coordinated activity of cytosolic and mitochondrial metabolic pathways to provide ATP and building blocks for DNA, RNA and protein synthesis. Many metabolic pathway genes are targets of the c-myc oncogene and cell-cycle regulator. However, the contribution of c-Myc to the activation of cytosolic and mitochondrial metabolic networks during cell-cycle entry is unknown. Here, we report the metabolic fates of [U-13C] glucose in serum-stimulated myc−/− and myc+/+ fibroblasts by 13C isotopomer NMR analysis. We demonstrate that endogenous c-myc increased 13C labeling of ribose sugars, purines and amino acids, indicating partitioning of glucose carbons into C1/folate and pentose phosphate pathways, and increased tricarboxylic acid cycle turnover at the expense of anaplerotic flux. Myc expression also increased global O-linked N-acetylglucosamine protein modification, and inhibition of hexosamine biosynthesis selectively reduced growth of Myc-expressing cells, suggesting its importance in Myc-induced proliferation. These data reveal a central organizing function for the Myc oncogene in the metabolism of cycling cells. The pervasive deregulation of this oncogene in human cancers may be explained by its function in directing metabolic networks required for cell proliferation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F . (2006). The c-Myc target gene network. Semin Cancer Biol 16: 253–264.
Dean M, Levine RA, Ran W, Kindy MS, Sonenshein GE, Campisi J . (1986). Regulation of c-myc transcription and mRNA abundance by serum growth factors and cell contact. J Biol Chem 261: 9161–9166.
Hart GW, Housley MP, Slawson C . (2007). Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446: 1017–1022.
Hunt AN, Postle AD . (2004). Phosphatidylcholine biosynthesis inside the nucleus: is it involved in regulating cell proliferation? Adv Enzyme Regul 44: 173–186.
Jackowski S . (1994). Coordination of membrane phospholipid synthesis with the cell cycle. J Biol Chem 269: 3858–3867.
Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN . (2006). Myc influences global chromatin structure. EMBO J 25: 2723–2734.
Lawlor ER, Soucek L, Brown-Swigart L, Shchors K, Bialucha CU, Evan GI . (2006). Reversible kinetic analysis of Myc targets in vivo provides novel insights into Myc-mediated tumorigenesis. Cancer Res 66: 4591–4601.
Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O’Donnell KA et al. (2005). Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol 25: 6225–6234.
Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B . (2003). A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc Natl Acad Sci USA 100: 8164–8169.
Lum JJ, Bui T, Gruber M, Gordan JD, DeBerardinis RJ, Covello KL et al. (2007). The transcription factor HIF-1alpha plays a critical role in the growth factor-dependent regulation of both anaerobic and aerobic glycolysis. Genes Dev 21: 1037–1049.
Malloy CR, Sherry AD, Jeffrey FM . (1990). Analysis of tricarboxylic acid cycle of the heart using 13C isotope isomers. Am J Physiol 259: H987–H995.
Mandal S, Guptan P, Owusu-Ansah E, Banerjee U . (2005). Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev Cell 9: 843–854.
Mannava S, Grachtchouk V, Wheeler LJ, Im M, Zhuang D, Slavina EG et al. (2008). Direct role of nucleotide metabolism in C-MYC-dependent proliferation of melanoma cells. Cell Cycle 7: 2392–2400.
Morrish FM, Neretti N, Sedivy JM, Hockenbery DM . (2008). The oncogene c-Myc coordinates regulation of metabolic networks to enable rapid cell cycle entry. Cell Cycle 7: 1054–1066.
Nikiforov MA, Chandriani S, O’Connell B, Petrenko O, Kotenko I, Beavis A et al. (2002). A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol Cell Biol 22: 5793–5800.
Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M et al. (2000). Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem 275: 21797–21800.
Peet AC, McConville C, Wilson M, Levine BA, Reed M, Dyer SA et al. (2007). 1H MRS identifies specific metabolite profiles associated with MYCN-amplified and non-amplified tumour subtypes of neuroblastoma cell lines. NMR Biomed 20: 692–700.
Schorl C, Sedivy JM . (2003). Loss of protooncogene c-Myc function impedes G1 phase progression both before and after the restriction point. Mol Biol Cell 14: 823–835.
Slawson C, Zachara NE, Vosseller K, Cheung WD, Lane MD, Hart GW . (2005). Perturbations in O-linked beta-N-acetylglucosamine protein modification cause severe defects in mitotic progression and cytokinesis. J Biol Chem 280: 32944–32956.
Tollefsbol TO, Cohen HJ . (1990). The protein synthetic surge in response to mitogen triggers high glycolytic enzyme levels in human lymphocytes and occurs prior to DNA synthesis. Biochem Med Metab Biol 44: 282–291.
Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT et al. (2006). Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci USA 103: 17834–17839.
Acknowledgements
We thank John Sedivy for cell lines. This work utilized the MMC database supported by NIH grants R21 DK070297 and P41 RR02301, the MDL database (www.mdl.imb.liu.se) and the Human Metabolome database (www.hmbd.ca). A portion of this research was performed at EMSL, a national scientific user facility sponsored by the Department of Energy's Office of Biological and Environmental Research at Pacific Northwest National Laboratory. This work was funded by RO1CA106650-02 (DH). Development of the program tcaCALC (University of Texas Southwestern Medical Center) was supported by H47669-16, a Department of Veterans Affairs Merit Review Award to CR Malloy, and RR02584.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
Supplementary information
Rights and permissions
About this article
Cite this article
Morrish, F., Isern, N., Sadilek, M. et al. c-Myc activates multiple metabolic networks to generate substrates for cell-cycle entry. Oncogene 28, 2485–2491 (2009). https://doi.org/10.1038/onc.2009.112
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2009.112
Keywords
This article is cited by
-
MYC sensitises cells to apoptosis by driving energetic demand
Nature Communications (2022)
-
MEN1 silencing aggravates tumorigenic potential of AR-independent prostate cancer cells through nuclear translocation and activation of JunD and β-catenin
Journal of Experimental & Clinical Cancer Research (2021)
-
USP16 regulates castration-resistant prostate cancer cell proliferation by deubiquitinating and stablizing c-Myc
Journal of Experimental & Clinical Cancer Research (2021)
-
Mitochondria transfer enhances proliferation, migration, and osteogenic differentiation of bone marrow mesenchymal stem cell and promotes bone defect healing
Stem Cell Research & Therapy (2020)
-
Regulation of cancer cell metabolism: oncogenic MYC in the driver’s seat
Signal Transduction and Targeted Therapy (2020)