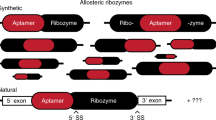
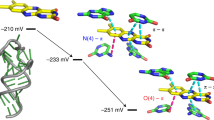
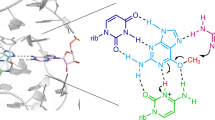
Abstract
Evolution has resoundingly favored protein enzymes over RNA-based catalysts, yet ribozymes occupy important niches in modern cell biology that include the starring role in catalysis of protein synthesis on the ribosome. Recent results from structural and biochemical studies show that natural ribozymes use an impressive range of catalytic mechanisms, beyond metalloenzyme chemistry and analogous to more chemically diverse protein enzymes. These findings make it increasingly possible to compare details of RNA- and protein-based catalysis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Jencks, W.P. Catalysis in Chemistry and Enzymology, 864 (Dover Publications, New York, 1987).
Narlikar, G.J. & Herschlag, D. Mechanistic aspects of enzymatic catalysis: lessons from comparison of RNA and protein enzymes. Annu. Rev. Biochem. 66, 19–59 (1997).
Armstrong, A.A. & Amzel, L.M. Role of entropy in increased rates of intramolecular reactions. J. Am. Chem. Soc. 125, 14596–14602 (2003).
Fersht, A.R. Catalysis, binding and enzyme-substrate complementarity. Proc. R. Soc. Lond. B. Biol. Sci. 187, 397–407 (1974).
Jencks, W.P. Imidazole and proton transfer in catalysis. Biochem. Soc. Symp. 31, 59–80 (1970).
Tesmer, J.J. et al. Two-metal-ion catalysis in adenylyl cyclase. Science 285, 756–760 (1999).
Wyckoff, H.W. et al. The three-dimensional structure of ribonuclease-S. Interpretation of an electron density map at a nominal resolution of 2 Å. J. Biol. Chem. 245, 305–328 (1970).
Drum, C.L. et al. Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin. Nature 415, 396–402 (2002).
Tang, J. & Breaker, R.R. Structural diversity of self-cleaving ribozymes. Proc. Natl. Acad. Sci. USA 97, 5784–5789 (2000).
Salehi-Ashtiani, K. & Szostak, J.W. In vitro evolution suggests multiple origins for the hammerhead ribozyme. Nature 414, 82–84 (2001).
Khvorova, A., Lescoute, A., Westhof, E. & Jayasena, S.D. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Biol. 10, 708–712 (2003).
Canny, M.D. et al. Fast cleavage kinetics of a natural hammerhead ribozyme. J. Am. Chem. Soc. 126, 10848–10849 (2004).
Penedo, J.C., Wilson, T.J., Jayasena, S.D., Khvorova, A. & Lilley, D.M. Folding of the natural hammerhead ribozyme is enhanced by interaction of auxiliary elements. RNA 10, 880–888 (2004).
Stage-Zimmermann, T.K. & Uhlenbeck, O.C. Hammerhead ribozyme kinetics. RNA 4, 875–889 (1998).
Raines, R.T. Active site of ribonuclease A. In Nucleic Acids and Molecular Biology Vol. 13 (ed. Zenkova, M.A.) (Springer, Heidelberg, Germany, 2004).
Murray, J.B., Seyhan, A.A., Walter, N.G., Burke, J.M. & Scott, W.G. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol. 5, 587–595 (1998).
Curtis, E.A. & Bartel, D.P. The hammerhead cleavage reaction in monovalent cations. RNA 7, 546–552 (2001).
O'Rear, J.L. et al. Comparison of the hammerhead cleavage reactions stimulated by monovalent and divalent cations. RNA 7, 537–545 (2001).
Pley, H.W., Flaherty, K.M. & McKay, D.B. Three-dimensional structure of a hammerhead ribozyme. Nature 372, 68–74 (1994).
Scott, W.G., Finch, J.T. & Klug, A. The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell 81, 991–1002 (1995).
Scott, W.G., Murray, J.B., Arnold, J.R., Stoddard, B.L. & Klug, A. Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme. Science 274, 2065–2069 (1996).
Murray, J.B. et al. The structural basis of hammerhead ribozyme self-cleavage. Cell 92, 665–673 (1998).
Murray, J.B., Szoke, H., Szoke, A. & Scott, W.G. Capture and visualization of a catalytic RNA enzyme-product complex using crystal lattice trapping and X-ray holographic reconstruction. Mol. Cell 5, 279–287 (2000).
Murray, J.B., Dunham, C.M. & Scott, W.G. A pH-dependent conformational change, rather than the chemical step, appears to be rate-limiting in the hammerhead ribozyme cleavage reaction. J. Mol. Biol. 315, 121–130 (2002).
Dunham, C.M., Murray, J.B. & Scott, W.G. A helical twist-induced conformational switch activates cleavage in the hammerhead ribozyme. J. Mol. Biol. 332, 327–336 (2003).
Ruffner, D.E. & Uhlenbeck, O.C. Thiophosphate interference experiments locate phosphates important for the hammerhead RNA self-cleavage reaction. Nucleic Acids Res. 18, 6025–6029 (1990).
Peracchi, A., Beigelman, L., Scott, E.C., Uhlenbeck, O.C. & Herschlag, D. Involvement of a specific metal ion in the transition of the hammerhead ribozyme to its catalytic conformation. J. Biol. Chem. 272, 26822–26826 (1997).
Scott, E.C. & Uhlenbeck, O.C. A re-investigation of the thio effect at the hammerhead cleavage site. Nucleic Acids Res. 27, 479–484 (1999).
Wang, S., Karbstein, K., Peracchi, A., Beigelman, L. & Herschlag, D. Identification of the hammerhead ribozyme metal ion binding site responsible for rescue of the deleterious effect of a cleavage site phosphorothioate. Biochemistry 38, 14363–14378 (1999).
Murray, J.B. & Scott, W.G. Does a single metal ion bridge the A-9 and scissile phosphate groups in the catalytically active hammerhead ribozyme structure? J. Mol. Biol. 296, 33–41 (2000).
Ferre-D'Amare, A.R., Zhou, K. & Doudna, J.A. Crystal structure of a hepatitis δ virus ribozyme. Nature 395, 567–574 (1998).
Rupert, P.B. & Ferre-D'Amare, A.R. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature 410, 780–786 (2001).
Bevilacqua, P.C. Mechanistic considerations for general acid-base catalysis by RNA: revisiting the mechanism of the hairpin ribozyme. Biochemistry 42, 2259–2265 (2003).
Rajagopal, P. & Feigon, J. Triple-strand formation in the homopurine:homopyrimidine DNA oligonucleotides d(G-A)4 and d(T-C)4. Nature 339, 637–640 (1989).
Sklenar, V. & Feigon, J. Formation of a stable triplex from a single DNA strand. Nature 345, 836–838 (1990).
Connell, G.J. & Yarus, M. RNAs with dual specificity and dual RNAs with similar specificity. Science 264, 1137–1141 (1994).
Legault, P. & Pardi, A. In situ probing of adenine protonation in RNA by 13C NMR. J. Am. Chem. Soc. 116, 8390–8391 (1994).
Ravindranathan, S., Butcher, S.E. & Feigon, J. Adenine protonation in domain B of the hairpin ribozyme. Biochemistry 39, 16026–16032 (2000).
Been, M.D. & Wickham, G.S. Self-cleaving ribozymes of hepatitis δ virus RNA. Eur. J. Biochem. 247, 741–753 (1997).
Thompson, J.E., Venegas, F.D. & Raines, R.T. Energetics of catalysis by ribonucleases: fate of the 2′,3′-cyclic phosphodiester intermediate. Biochemistry 33, 7408–7414 (1994).
Lebruska, L.L., Kuzmine, II & Fedor, M.J. Rescue of an abasic hairpin ribozyme by cationic nucleobases: evidence for a novel mechanism of RNA catalysis. Chem. Biol. 9, 465–473 (2002).
Perrotta, A.T., Shih, I. & Been, M.D. Imidazole rescue of a cytosine mutation in a self-cleaving ribozyme. Science 286, 123–126 (1999).
Shih, I.H. & Been, M.D. Involvement of a cytosine side chain in proton transfer in the rate-determining step of ribozyme self-cleavage. Proc. Natl. Acad. Sci. USA 98, 1489–1494 (2001).
Nakano, S., Chadalavada, D.M. & Bevilacqua, P.C. General acid-base catalysis in the mechanism of a hepatitis δ virus ribozyme. Science 287, 1493–1497 (2000).
Nakano, S., Proctor, D.J. & Bevilacqua, P.C. Mechanistic characterization of the HDV genomic ribozyme: assessing the catalytic and structural contributions of divalent metal ions within a multichannel reaction mechanism. Biochemistry 40, 12022–12038 (2001).
Nakano, S. & Bevilacqua, P.C. Proton inventory of the genomic HDV ribozyme in Mg(2+)-containing solutions. J. Am. Chem. Soc. 123, 11333–11334 (2001).
Luptak, A., Ferre-D'Amare, A.R., Zhou, K., Zilm, K.W. & Doudna, J.A. Direct pK(a) measurement of the active-site cytosine in a genomic hepatitis δ virus ribozyme. J. Am. Chem. Soc. 123, 8447–8452 (2001).
Ke, A., Zhou, K., Ding, F., Cate, J.H. & Doudna, J.A. A conformational switch controls hepatitis δ virus ribozyme catalysis. Nature 429, 201–205 (2004).
Kuzmin, Y.I., Da Costa, C.P. & Fedor, M.J. Role of an active site guanine in hairpin ribozyme catalysis probed by exogenous nucleobase rescue. J. Mol. Biol. 340, 233–251 (2004).
Nahas, M.K. et al. Observation of internal cleavage and ligation reactions of a ribozyme. Nat. Struct. Mol. Biol. 11, 1107–1113 (2004).
Rupert, P.B., Massey, A.P., Sigurdsson, S.T. & Ferre-D'Amare, A.R. Transition state stabilization by a catalytic RNA. Science 298, 1421–1424 (2002).
Ryder, S.P. et al. Investigation of adenosine base ionization in the hairpin ribozyme by nucleotide analog interference mapping. RNA 7, 1454–1463 (2001).
Hampel, A. & Cowan, J.A. A unique mechanism for RNA catalysis: the role of metal cofactors in hairpin ribozyme cleavage. Chem. Biol. 4, 513–517 (1997).
Nesbitt, S., Hegg, L.A. & Fedor, M.J. An unusual pH-independent and metal-ion-independent mechanism for hairpin ribozyme catalysis. Chem. Biol. 4, 619–630 (1997).
Hampel, K.J. & Burke, J.M. A conformational change in the “loop E-like” motif of the hairpin ribozyme is coincidental with domain docking and is essential for catalysis. Biochemistry 40, 3723–3729 (2001).
Blount, K.F., Grover, N.L., Mokler, V., Beigelman, L. & Uhlenbeck, O.C. Steric interference modification of the hammerhead ribozyme. Chem. Biol. 9, 1009–1016 (2002).
Pereira, M.J., Harris, D.A., Rueda, D. & Walter, N.G. Reaction pathway of the trans-acting hepatitis δ virus ribozyme: a conformational change accompanies catalysis. Biochemistry 41, 730–740 (2002).
Hampel, K.J. & Burke, J.M. Solvent protection of the hammerhead ribozyme in the ground state: evidence for a cation-assisted conformational change leading to catalysis. Biochemistry 42, 4421–4429 (2003).
Doherty, E.A., Herschlag, D. & Doudna, J.A. Assembly of an exceptionally stable RNA tertiary interface in a group I ribozyme. Biochemistry 38, 2982–2990 (1999).
Engelhardt, M.A., Doherty, E.A., Knitt, D.S., Doudna, J.A. & Herschlag, D. The P5abc peripheral element facilitates preorganization of the Tetrahymena group I ribozyme for catalysis. Biochemistry 39, 2639–2651 (2000).
Ohuchi, S.J., Ikawa, Y., Shiraishi, H. & Inoue, T. Modular engineering of a Group I intron ribozyme. Nucleic Acids Res. 30, 3473–3480 (2002).
De la Pena, M., Gago, S. & Flores, R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 22, 5561–5570 (2003).
Costa, M., Michel, F. & Westhof, E. A three-dimensional perspective on exon binding by a group II self-splicing intron. EMBO J. 19, 5007–5018 (2000).
Swisher, J., Duarte, C.M., Su, L.J. & Pyle, A.M. Visualizing the solvent-inaccessible core of a group II intron ribozyme. EMBO J. 20, 2051–2061 (2001).
Zhang, L. & Doudna, J.A. Structural insights into group II intron catalysis and branch-site selection. Science 295, 2084–2088 (2002).
Sigel, R.K. et al. Solution structure of domain 5 of a group II intron ribozyme reveals a new RNA motif. Nat. Struct. Mol. Biol. 11, 187–192 (2004).
Herschlag, D. & Cech, T.R. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry 29, 10159–10171 (1990).
Herschlag, D. & Cech, T.R. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 2. Kinetic description of the reaction of an RNA substrate that forms a mismatch at the active site. Biochemistry 29, 10172–10180 (1990).
McSwiggen, J.A. & Cech, T.R. Stereochemistry of RNA cleavage by the Tetrahymena ribozyme and evidence that the chemical step is not rate-limiting. Science 244, 679–683 (1989).
Rajagopal, J., Doudna, J.A. & Szostak, J.W. Stereochemical course of catalysis by the Tetrahymena ribozyme. Science 244, 692–694 (1989).
Shan, S., Yoshida, A., Sun, S., Piccirilli, J.A. & Herschlag, D. Three metal ions at the active site of the Tetrahymena group I ribozyme. Proc. Natl. Acad. Sci. USA 96, 12299–12304 (1999).
Shan, S.O. & Herschlag, D. Probing the role of metal ions in RNA catalysis: kinetic and thermodynamic characterization of a metal ion interaction with the 2′-moiety of the guanosine nucleophile in the Tetrahymena group I ribozyme. Biochemistry 38, 10958–10975 (1999).
Shan, S., Kravchuk, A.V., Piccirilli, J.A. & Herschlag, D. Defining the catalytic metal ion interactions in the Tetrahymena ribozyme reaction. Biochemistry 40, 5161–5171 (2001).
Piccirilli, J.A., Vyle, J.S., Caruthers, M.H. & Cech, T.R. Metal ion catalysis in the Tetrahymena ribozyme reaction. Nature 361, 85–88 (1993).
Narlikar, G.J., Gopalakrishnan, V., McConnell, T.S., Usman, N. & Herschlag, D. Use of binding energy by an RNA enzyme for catalysis by positioning and substrate destabilization. Proc. Natl. Acad. Sci. USA 92, 3668–3672 (1995).
Cate, J.H. et al. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science 273, 1678–1685 (1996).
Lehnert, V., Jaeger, L., Michel, F. & Westhof, E. New loop-loop tertiary interactions in self-splicing introns of subgroup IC and ID: a complete 3D model of the Tetrahymena thermophila ribozyme. Chem. Biol. 3, 993–1009 (1996).
Golden, B.L., Gooding, A.R., Podell, E.R. & Cech, T.R. A preorganized active site in the crystal structure of the Tetrahymena ribozyme. Science 282, 259–264 (1998).
Adams, P.L., Stahley, M.R., Kosek, A.B., Wang, J. & Strobel, S.A. Crystal structure of a self-splicing group I intron with both exons. Nature 430, 45–50 (2004).
Guo, F., Gooding, A.R. & Cech, T.R. Structure of the Tetrahymena ribozyme: base triple sandwich and metal ion at the active site. Mol. Cell 16, 351–362 (2004).
Golden, B.L., Kim, H. & Chase, E. Crystal structure of a phage Twort group I ribozyme–product complex. Nat. Struct. Mol. Biol. 12, 82–89 (2005).
Steitz, T.A. & Steitz, J.A. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. USA 90, 6498–6502 (1993).
Vortler, L.C. & Eckstein, F. Phosphorothioate modification of RNA for stereochemical and interference analyses. Methods Enzymol. 317, 74–91 (2000).
Keating, T.A. & Walsh, C.T. Initiation, elongation, and termination strategies in polyketide and polypeptide antibiotic biosynthesis. Curr. Opin. Chem. Biol. 3, 598–606 (1999).
Sieber, S.A. & Marahiel, M.A. Learning from nature's drug factories: nonribosomal synthesis of macrocyclic peptides. J. Bacteriol. 185, 7036–7043 (2003).
Walsh, C.T. Polyketide and nonribosomal peptide antibiotics: modularity and versatility. Science 303, 1805–1810 (2004).
Moazed, D. & Noller, H.F. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell 57, 585–597 (1989).
Noller, H.F., Hoffarth, V. & Zimniak, L. Unusual resistance of peptidyl transferase to protein extraction procedures. Science 256, 1416–1419 (1992).
Samaha, R.R., Green, R. & Noller, H.F. A base pair between tRNA and 23S rRNA in the peptidyl transferase centre of the ribosome. Nature 377, 309–314 (1995).
Green, R., Samaha, R.R. & Noller, H.F. Mutations at nucleotides G2251 and U2585 of 23 S rRNA perturb the peptidyl transferase center of the ribosome. J. Mol. Biol. 266, 40–50 (1997).
Green, R., Switzer, C. & Noller, H.F. Ribosome-catalyzed peptide-bond formation with an A-site substrate covalently linked to 23S ribosomal RNA. Science 280, 286–289 (1998).
Ban, N., Nissen, P., Hansen, J., Moore, P.B. & Steitz, T.A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289, 905–920 (2000).
Sievers, A., Beringer, M., Rodnina, M.V. & Wolfenden, R. The ribosome as an entropy trap. Proc. Natl. Acad. Sci. USA 101, 7897–7901 (2004).
Nissen, P., Hansen, J., Ban, N., Moore, P.B. & Steitz, T.A. The structural basis of ribosome activity in peptide bond synthesis. Science 289, 920–930 (2000).
Muth, G.W., Ortoleva-Donnelly, L. & Strobel, S.A. A single adenosine with a neutral pKa in the ribosomal peptidyl transferase center. Science 289, 947–950 (2000).
Asai, T. et al. Construction and initial characterization of Escherichia coli strains with few or no intact chromosomal rRNA operons. J. Bacteriol. 181, 3803–3809 (1999).
Katunin, V.I., Muth, G.W., Strobel, S.A., Wintermeyer, W. & Rodnina, M.V. Important contribution to catalysis of peptide bond formation by a single ionizing group within the ribosome. Mol. Cell 10, 339–346 (2002).
Youngman, E.M., Brunelle, J.L., Kochaniak, A.B. & Green, R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 117, 589–599 (2004).
Pedersen, S. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J. 3, 2895–2898 (1984).
Weinger, J.S., Parnell, K.M., Dorner, S., Green, R. & Strobel, S.A. Substrate-assisted catalysis of peptide bond formation by the ribosome. Nat. Struct. Mol. Biol. 11, 1101–1106 (2004).
Winkler, W.C., Nahvi, A., Roth, A., Collins, J.A. & Breaker, R.R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428, 281–286 (2004).
Teixeira, A. et al. Autocatalytic RNA cleavage in the human β-globin pre-mRNA promotes transcription termination. Nature 432, 526–530 (2004).
Xu, R.X. et al. Atomic structure of PDE4: insights into phosphodiesterase mechanism and specificity. Science 288, 1822–1825 (2000).
Mildvan, A.S. et al. Structures and mechanisms of Nudix hydrolases. Arch. Biochem. Biophys. 433, 129–143 (2005).
Huai, Q., Colicelli, J. & Ke, H. The crystal structure of AMP-bound PDE4 suggests a mechanism for phosphodiesterase catalysis. Biochemistry 42, 13220–13226 (2003).
Schmeing, T.M. et al. A pre-translocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nat. Struct. Biol. 9, 225–230 (2002).
Acknowledgements
The authors thank E. Friedman, K. Karbstein and A. Ke for helpful comments on the manuscript, R. Green and A. Mildvan for stimulating discussions, and A. Ke and S. Dorner for preparation of Figures 1b,d, 2b,d and 3b.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Doudna, J., Lorsch, J. Ribozyme catalysis: not different, just worse. Nat Struct Mol Biol 12, 395–402 (2005). https://doi.org/10.1038/nsmb932
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb932
This article is cited by
-
Biotechnological and Therapeutic Applications of Natural Nucleic Acid Structural Motifs
Topics in Current Chemistry (2020)
-
Development of a ribonuclease containing a G4-specific binding motif for programmable RNA cleavage
Scientific Reports (2019)
-
The structural stability and catalytic activity of DNA and RNA oligonucleotides in the presence of organic solvents
Biophysical Reviews (2016)
-
Does the Ribosome Challenge our Understanding of the RNA World?
Journal of Molecular Evolution (2016)
-
Are Molecular Alphabets Universal Enabling Factors for the Evolution of Complex Life?
Origins of Life and Evolution of Biospheres (2013)