Abstract
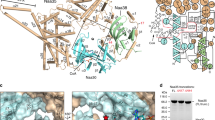
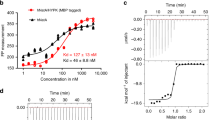
N-terminal acetylation is ubiquitous among eukaryotic proteins and controls a myriad of biological processes. Of the N-terminal acetyltransferases (NATs) that facilitate this cotranslational modification, the heterodimeric NatA complex has the most diversity for substrate selection and modifies the majority of all N-terminally acetylated proteins. Here, we report the X-ray crystal structure of the 100-kDa holo-NatA complex from Schizosaccharomyces pombe, in the absence and presence of a bisubstrate peptide-CoA–conjugate inhibitor, as well as the structure of the uncomplexed Naa10p catalytic subunit. The NatA-Naa15p auxiliary subunit contains 13 tetratricopeptide motifs and adopts a ring-like topology that wraps around the NatA-Naa10p subunit, an interaction that alters the Naa10p active site for substrate-specific acetylation. These studies have implications for understanding the mechanistic details of other NAT complexes and how regulatory subunits modulate the activity of the broader family of protein acetyltransferases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Arnesen, T. et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc. Natl. Acad. Sci. USA 106, 8157–8162 (2009).
Forte, G.M., Pool, M.R. & Stirling, C.J. N-terminal acetylation inhibits protein targeting to the endoplasmic reticulum. PLoS Biol. 9, e1001073 (2011).
Hwang, C.S., Shemorry, A. & Varshavsky, A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science 327, 973–977 (2010).
Scott, D.C., Monda, J.K., Bennett, E.J., Harper, J.W. & Schulman, B.A. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science 334, 674–678 (2011).
Starheim, K.K., Gevaert, K. & Arnesen, T. Protein N-terminal acetyltransferases: when the start matters. Trends Biochem. Sci. 37, 152–161 (2012).
Yi, C.H. et al. Metabolic regulation of protein N-alpha-acetylation by Bcl-xL promotes cell survival. Cell 146, 607–620 (2011).
Arnesen, T. et al. Identification and characterization of the human ARD1-NATH protein acetyltransferase complex. Biochem. J. 386, 433–443 (2005).
Gautschi, M. et al. The yeast Nα-acetyltransferase NatA is quantitatively anchored to the ribosome and interacts with nascent polypeptides. Mol. Cell Biol. 23, 7403–7414 (2003).
Mullen, J.R. et al. Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J. 8, 2067–2075 (1989).
Park, E.C. & Szostak, J.W. ARD1 and NAT1 proteins form a complex that has N-terminal acetyltransferase activity. EMBO J. 11, 2087–2093 (1992).
Polevoda, B., Brown, S., Cardillo, T.S., Rigby, S. & Sherman, F. Yeast Nα-terminal acetyltransferases are associated with ribosomes. J. Cell Biochem. 103, 492–508 (2008).
Starheim, K.K. et al. Identification of the human Nα-acetyltransferase complex B (hNatB): a complex important for cell-cycle progression. Biochem. J. 415, 325–331 (2008).
Starheim, K.K. et al. Knockdown of human Nα-terminal acetyltransferase complex C leads to p53-dependent apoptosis and aberrant human Arl8b localization. Mol. Cell Biol. 29, 3569–3581 (2009).
Arnesen, T., Thompson, P.R., Varhaug, J.E. & Lillehaug, J.R. The protein acetyltransferase ARD1: a novel cancer drug target? Curr. Cancer Drug Targets 8, 545–553 (2008).
Arnesen, T. et al. Induction of apoptosis in human cells by RNAi-mediated knockdown of hARD1 and NATH, components of the protein N-α-acetyltransferase complex. Oncogene 25, 4350–4360 (2006).
Lim, J.H., Park, J.W. & Chun, Y.S. Human arrest defective 1 acetylates and activates β-catenin, promoting lung cancer cell proliferation. Cancer Res. 66, 10677–10682 (2006).
Ren, T. et al. Generation of novel monoclonal antibodies and their application for detecting ARD1 expression in colorectal cancer. Cancer Lett. 264, 83–92 (2008).
Kalvik, T.V. & Arnesen, T. Protein N-terminal acetyltransferases in cancer. Oncogene 32, 269–276 (2013).
Yu, M. et al. Correlation of expression of human arrest-defective-1 (hARD1) protein with breast cancer. Cancer Invest. 27, 978–983 (2009).
Fluge, Ø., Bruland, O., Akslen, L.A., Varhaug, J.E. & Lillehaug, J.R. NATH, a novel gene overexpressed in papillary thyroid carcinomas. Oncogene 21, 5056–5068 (2002).
Yu, M. et al. Immunohistochemical analysis of human arrest-defective-1 expressed in cancers in vivo. Oncol. Rep. 21, 909–915 (2009).
Polevoda, B. & Sherman, F. Composition and function of the eukaryotic N-terminal acetyltransferase subunits. Biochem. Biophys. Res. Commun. 308, 1–11 (2003).
Polevoda, B. & Sherman, F. N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J. Mol. Biol. 325, 595–622 (2003).
Starheim, K.K., Gromyko, D., Velde, R., Varhaug, J.E. & Arnesen, T. Composition and biological significance of the human Nα-terminal acetyltransferases. BMC Proc. 3 (suppl. 6), S3 (2009).
Polevoda, B., Norbeck, J., Takakura, H., Blomberg, A. & Sherman, F. Identification and specificities of N-terminal acetyltransferases from Saccharomyces cerevisiae. EMBO J. 18, 6155–6168 (1999).
Van Damme, P. et al. Proteome-derived peptide libraries allow detailed analysis of the substrate specificities of Nα-acetyltransferases and point to hNaa10p as the post-translational actin Nα-acetyltransferase. Mol. Cell Proteomics 10, M110.004580 (2011).
Van Damme, P. et al. N-terminal acetylome analyses and functional insights of the N-terminal acetyltransferase NatB. Proc. Natl. Acad. Sci. USA 109, 12449–12454 (2012).
Sutton, A. et al. Sas4 and Sas5 are required for the histone acetyltransferase activity of Sas2 in the SAS complex. J. Biol. Chem. 278, 16887–16892 (2003).
Iizuka, M. & Stillman, B. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J. Biol. Chem. 274, 23027–23034 (1999).
Parthun, M.R., Widom, J. & Gottschling, D.E. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell 87, 85–94 (1996).
Polevoda, B., Hoskins, J. & Sherman, F. Properties of Nat4, an Nα-acetyltransferase of Saccharomyces cerevisiae that modifies N termini of histones H2A and H4. Mol. Cell Biol. 29, 2913–2924 (2009).
Hole, K. et al. The human N-alpha-acetyltransferase 40 (hNaa40p/hNatD) is conserved from yeast and N-terminally acetylates histones H2A and H4. PLoS ONE 6, e24713 (2011).
Van Damme, P. et al. NatF contributes to an evolutionary shift in protein N-terminal acetylation and is important for normal chromosome segregation. PLoS Genet. 7, e1002169 (2011).
Song, O.K., Wang, X., Waterborg, J.H. & Sternglanz, R. An Nα-acetyltransferase responsible for acetylation of the N-terminal residues of histones H4 and H2A. J. Biol. Chem. 278, 38109–38112 (2003).
Evjenth, R. et al. Human Naa50p (Nat5/San) displays both protein Nα- and Nå-acetyltransferase activity. J. Biol. Chem. 284, 31122–31129 (2009).
Liszczak, G., Arnesen, T. & Marmorstein, R. Structure of a ternary Naa50p (NAT5/SAN) N-terminal acetyltransferase complex reveals the molecular basis for substrate-specific acetylation. J. Biol. Chem. 286, 37002–37010 (2011).
Blatch, G.L. & Lassle, M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21, 932–939 (1999).
Arnesen, T. et al. The chaperone-like protein HYPK acts together with NatA in cotranslational N-terminal acetylation and prevention of Huntingtin aggregation. Mol. Cell Biol. 30, 1898–1909 (2010).
Cingolani, G., Petosa, C., Weis, K. & Muller, C.W. Structure of importin-β bound to the IBB domain of importin-α. Nature 399, 221–229 (1999).
Vetting, M.W. et al. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 433, 212–226 (2005).
Tanner, K.G. et al. Catalytic mechanism and function of invariant glutamic acid 173 from the histone acetyltransferase GCN5 transcriptional coactivator. J. Biol. Chem. 274, 18157–18160 (1999).
Berndsen, C.E., Albaugh, B.N., Tan, S. & Denu, J.M. Catalytic mechanism of a MYST family histone acetyltransferase. Biochemistry 46, 623–629 (2007).
Evjenth, R.H. et al. Human protein N-terminal acetyltransferase hNaa50p (hNAT5/hSAN) follows ordered sequential catalytic mechanism: combined kinetic and NMR study. J. Biol. Chem. 287, 10081–10088 (2012).
Braman, J., Papworth, C. & Greener, A. Site-directed mutagenesis using double-stranded plasmid DNA templates. Methods Mol. Biol. 57, 31–44 (1996).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Langer, G., Cohen, S.X., Lamzin, V.S. & Perrakis, A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 (2008).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Painter, J. & Merritt, E.A. A molecular viewer for the analysis of TLS rigid-body motion in macromolecules. Acta Crystallogr. D Biol. Crystallogr. 61, 465–471 (2005).
Painter, J. & Merritt, E.A. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 (2006).
Terwilliger, T.C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 (1999).
Terwilliger, T.C. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 (2000).
Acknowledgements
This work was supported by US National Institutes of Health (NIH) grant GM060293 (R.M.) and NIH training grant GM071339 (G.L.). We acknowledge the use of the Wistar Proteomics Core facility for the work reported here, which is supported in part by NIH grant CA010815. T.A. was supported by the Research Council of Norway and the Norwegian Cancer Society. We also acknowledge Marmorstein laboratory members and E. Skordalakes for helpful discussions.
Author information
Authors and Affiliations
Contributions
G.L. performed all of the structural and biochemical experiments described in the manuscript, and J.M.G. carried out inhibitor synthesis. G.L. prepared manuscript figures, text and videos; H.F. carried out preliminary inhibition studies that led to experiments reported in the manuscript; R.M. designed and supervised experiments by G.L. and prepared manuscript text. T.A. supervised the experiments of H.F. and prepared manuscript text. E.J.P. supervised the experiments of J.M.G. All authors read and approved the submitted manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 (PDF 12310 kb)
An overall view of the NatA complex structure
The view shown in Fig. 1A is shown rotating 360° on the y-axis followed by 360° on the x-axis. (AVI 17786 kb)
Global conformational shifts in Naa10p upon Naa15p binding
This video begins with the uncomplexed Naa10p and highlights all residues featured in Figures 1c,d. It first shows the position of these residues in the uncomplexed Naa10p and a morph shows the change in position of these residues upon Naa15p binding. (AVI 29209 kb)
Naa10p active site conformational changes upon Naa15p binding
This movie shows a morph that highlights the change in position of Naa10p residues Leu22, Glu24, Tyr26, Arg113 and Tyr139 upon Naa15p binding. The morph is shown twice, once with AcCoA bound to Naa10p and again with the bisubstrate inhibitor-bound Naa10p. (AVI 12648 kb)
Rights and permissions
About this article
Cite this article
Liszczak, G., Goldberg, J., Foyn, H. et al. Molecular basis for N-terminal acetylation by the heterodimeric NatA complex. Nat Struct Mol Biol 20, 1098–1105 (2013). https://doi.org/10.1038/nsmb.2636
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2636
This article is cited by
-
NAA60 (HAT4): the newly discovered bi-functional Golgi member of the acetyltransferase family
Clinical Epigenetics (2022)
-
Naa10p promotes cell invasiveness of esophageal cancer by coordinating the c-Myc and PAI1 regulatory axis
Cell Death & Disease (2022)
-
Biochemical analysis of novel NAA10 variants suggests distinct pathogenic mechanisms involving impaired protein N-terminal acetylation
Human Genetics (2022)
-
NAA10 p.(N101K) disrupts N-terminal acetyltransferase complex NatA and is associated with developmental delay and hemihypertrophy
European Journal of Human Genetics (2021)
-
Structural basis of Naa20 activity towards a canonical NatB substrate
Communications Biology (2021)