Key Points
-
There are six fungal prions: four are self-propagating amyloids and two are self-activating enzymes.
-
[URE3] is a prion of the nitrogen catabolism regulator Ure2p; [PSI+] is a prion of the translation-termination factor Sup35p; [PIN+] is a prion of Rnq1p (function unknown); [Het-s] is a prion of the heterokaryon incompatibility protein HETs; [β] is a prion of vacuolar protease B; and [C] is a prion of a mitogen-activated protein kinase kinase kinase.
-
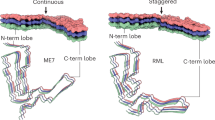
The infectious amyloid of Sup35p has a parallel in-register β-sheet structure.
-
The [Het-s] prion of Podospora anserina apparently benefits its host, but the [URE3] and [PSI+] prions of Saccharomyces cerevisiae are detrimental.
-
Chaperones catalyse amyloid filament breakage to form new seeds, and probably have other roles in prion propagation and generation as well.
-
Different prion variants, with the same protein sequence, have different amyloid structures. Variants can determine host range and chaperone effects.
Abstract
The term 'prion' means an infectious protein that does not need an accompanying nucleic acid. There are six fungal prions, including four self-propagating amyloids and two enzymes that are necessary to activate their inactive precursors. Here we explore the scope of the prion phenomenon, the biological and evolutionary roles of prions, the structural basis of the amyloid prions and the prominent role of chaperones (proteins that affect the folding of other proteins) and other cellular components in prion generation and propagation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
M'Gowan, J. P. Investigation into the disease of sheep called 'scrapie' (Blackwood, Edinburgh, 1914).
Wickner, R. B. Scrapie in ancient China? Science 309, 874 (2005).
Prusiner, S. B. (ed.) Prion Biology and Diseases (Cold Spring Harbor Laboratory Press, New York, 2004).
Chesebro, B. Introduction to the transmissible spongiform encephalopathies or prion diseases. Br. Med. Bull. 66, 1–20 (2003).
Wickner, R. B. [URE3] as an altered URE2 protein: evidence for a prion analog in S. cerevisiae. Science 264, 566–569 (1994). The original description of yeast prions, including the genetic criteria that distinguish prions from nucleic-acid replicons.
Wickner, R. B. in Fields Virology 5th edn (eds Knipe, D. M. & Howley, P. M.) 737–768 (Lippincott, Williams & Wilkins, 2006).
Cox, B. S. PSI, a cytoplasmic suppressor of super-suppressor in yeast. Heredity 20, 505–521 (1965).
Lacroute, F. Non-mendelian mutation allowing ureidosuccinic acid uptake in yeast. J. Bacteriol. 106, 519–522 (1971).
Cooper, T. G. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol. Revs. 26, 223–238 (2002).
Turoscy, V. & Cooper, T. G. Ureidosuccinate is transported by the allantoate transport system in Saccharomyces cerevisiae. J. Bacteriol. 169, 2598–2600 (1987).
Schlumpberger, M., Prusiner, S. B. & Herskowitz, I. Induction of distinct [URE3] yeast prion strains. Mol. Cell. Biol. 21, 7035–7046 (2001).
Brachmann, A., Baxa, U. & Wickner, R. B. Prion generation in vitro: amyloid of Ure2p is infectious. EMBO J. 24, 3082–3092 (2005).
Derkatch, I. L., Bradley, M. E., Zhou, P., Chernoff, Y. O. & Liebman, S. W. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 147, 507–519 (1997).
Derkatch, I. L., Bradley, M. E., Hong, J. Y. & Liebman, S. W. Prions affect the appearance of other prions: the story of [PIN]. Cell 106, 171–182 (2001). This report showed that Q/N-rich protein aggregates can prime [ PSI+] prion generation.
Sondheimer, N. & Lindquist, S. Rnq1: an epigenetic modifier of protein function in yeast. Molec. Cell 5, 163–172 (2000).
Rizet, G. Les phenomenes de barrage chez Podospora anserina: analyse genetique des barrages entre les souches s et S. Rev. Cytol. Biol. Veg. 13, 51–92 (1952).
Saupe, S. J. Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol. Mol. Biol. Revs 64, 489–502 (2000).
Benkemoun, L. & Saupe, S. J. Prion proteins as genetic material in fungi. Fungal Genet. Biol. 43, 789–803 (2006).
Coustou, V., Deleu, C., Saupe, S. & Begueret, J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc. Natl Acad. Sci. USA 94, 9773–9778 (1997). The original identification of [Het-s] as a prion.
Roberts, B. T. & Wickner, R. B. A class of prions that propagate via covalent auto-activation. Genes Dev. 17, 2083–2087 (2003).
Jones, E. W. Three proteolytic systems in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 266, 7963–7966 (1991).
Zubenko, G. S., Park, F. J. & Jones, E. W. Genetic properties of mutations at the PEP4 locus in Saccharomyces cerevisiae. Genetics 102, 679–690 (1982).
Kicka, S., Bonnet, C., Sobering, A. K., Ganesan, L. P. & Silar, P. A mitotically inheritable unit containing a MAP kinase module. Proc. Natl Acad. Sci. USA 103, 13445–13450 (2006).
Wickner, R. B. et al. Prions: proteins as genes and infectious entities. Genes Dev. 18, 470–485 (2004).
Maddelein, M. L., Dos Reis, S., Duvezin-Caubet, S., Coulary-Salin, B. & Saupe, S. J. Amyloid aggregates of the HET-s prion protein are infectious. Proc. Natl Acad. Sci. USA 99, 7402–7407 (2002). The first demonstrated transmission of a prion by an amyloid of recombinant protein.
King, C. Y. & Diaz-Avalos, R. Protein-only transmission of three yeast prion strains. Nature 428, 319–323 (2004).
Tanaka, M., Chien, P., Naber, N., Cooke, R. & Weissman, J. S. Conformational variations in an infectious protein determine prion strain differences. Nature 428, 323–328 (2004). References 26 and 27 showed that amyloid structure determines prion variant.
Patel, B. K. & Liebman, S. W. “Prion proof” for [PIN+]: infection with in vitro-made amyloid aggregates of Rnq1p-(132–405) induces [PIN+]. J. Mol. Biol. 365, 773–782 (2007).
Masison, D. C. & Wickner, R. B. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science 270, 93–95 (1995). The first biochemical evidence for yeast prions and prion domains.
Pierce, M. M., Baxa, U., Steven, A. C., Bax, A. & Wickner, R. B. Is the prion domain of soluble Ure2p unstructured? Biochemistry 44, 321–328 (2005).
TerAvanesyan, A., Dagkesamanskaya, A. R., Kushnirov, V. V. & Smirnov, V. N. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics 137, 671–676 (1994).
Doel, S. M., McCready, S. J., Nierras, C. R. & Cox, B. S. The dominant PNM2− mutation which eliminates the [PSI] factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics 137, 659–670 (1994).
DePace, A. H., Santoso, A., Hillner, P. & Weissman, J. S. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell 93, 1241–1252 (1998).
Mead, S. et al. Balancing selection at the prion protein gene consistent with prehistoric kurulike epidemics. Science 300, 640–643 (2003).
Kochneva-Pervukhova, N. V. et al. Mechanism of inhibition of Ψ+ prion determinant propagation by a mutation of the N-terminus of the yeast Sup35 protein. EMBO J. 17, 5805–5810 (1998).
Prusiner, S. B. et al. Transgenic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell 63, 673–686 (1990).
Priola, S. A., Caughey, B., Race, R. E. & Chesebro, B. Heterologous PrP molecules interfere with accumulation of protease-resistant PrP in scrapie-infected murine neuroblastoma cells. J. Virol. 68, 4873–4878 (1994).
Ross, E. D., Minton, A. P. & Wickner, R. B. Prion domains: sequences, structures and interactions. Nature Cell Biol. 7, 1039–1044 (2005).
Ross, E. D., Baxa, U. & Wickner, R. B. Scrambled prion domains form prions and amyloid. Mol. Cell. Biol. 24, 7206–7213 (2004).
Ross, E. D., Edskes, H. K., Terry, M. J. & Wickner, R. B. Primary sequence independence for prion formation. Proc. Natl Acad. Sci. USA 102, 12825–12830 (2005).
Chan, J. C. C., Oyler, N. A., Yau, W. M. & Tycko, R. Parallel β-sheets and polar zippers in amyloid fibrils formed by residues 10–39 of the yeast prion protein Ure2p. Biochemistry 44, 10669–10680 (2005).
Tycko, R. Molecular structure of amyloid fibrils: insights from solid-state NMR. Quart. Revs. Biophys. 1, 1–55 (2006).
Shewmaker, F., Wickner, R. B. & Tycko, R. Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc. Natl Acad. Sci. USA 103, 19754–19759 (2006). The first evidence-based prion amyloid structure.
Nelson, R. et al. Structure of the cross-β spine of amyloid-like fibrils. Nature 435, 773–778 (2005).
Krishnan, R. & Lindquist, S. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature 435, 765–772 (2005).
Diaz-Avalos, R., King, C. Y., Wall, J. S., Simon, M. & Caspar, D. L. D. Strain-specific morphologies of yeast prion amyloids. Proc. Natl Acad. Sci. USA 102, 10165–10170 (2005).
Balguerie, A. et al. Domain organization and structure–function relationship of the HET-s prion protein of Podospora anserina. EMBO J. 22, 2071–2081 (2003).
Ritter, C. et al. Correlation of structural elements and infectivity of the HET-s prion. Nature 435, 844–848 (2005).
Chernoff, Y. O. & Ono, B. I. in Protein Synthesis and Targeting in Yeast (eds Brown, A. J. P., Tuite, M. F. & McCarthy, J. E. G.) 101–107 (Springer, Berlin, 1992).
Chernoff, Y. O., Lindquist, S. L., Ono, B. I., Inge-Vechtomov, S. G. & Liebman, S. W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268, 880–884 (1995). The first demonstration of chaperone involvement in prion propagation.
Newnam, G. P., Wegrzyn, R. D., Lindquist, S. L. & Chernoff, Y. O. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol. Cell. Biol. 19, 1325–1333 (1999).
Jung, G., Jones, G., Wegrzyn, R. D. & Masison, D. C. A role for cytosolic Hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics 156, 559–570 (2000).
Kushnirov, V. V., Kryndushkin, D. S., Boguta, M., Smirnov, V. N. & Ter-Avanesyan, M. D. Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr. Biol. 10, 1443–1446 (2000).
Moriyama, H., Edskes, H. K. & Wickner, R. B. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol. Cell. Biol. 20, 8916–8922 (2000).
Jung, G. & Masison, D. C. Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr. Microbiol. 43, 7–10 (2001).
Sondheimer, N., Lopez, N., Craig, E. A. & Lindquist, S. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 20, 2435–2442 (2001).
Jones, G. W. & Masison, D. C. Saccharomyces cerevisiae Hsp70 mutations affect [PSI+] prion propagation and cell growth differently and implicate Hsp40 and tetratricopeptide repeat cochaperones in impairment of [PSI+]. Genetics 163, 495–506. (2003).
Jones, G., Song, Y., Chung, S. & Masison, D. C. Propagation of yeast [PSI+] prion impaired by factors that regulate Hsp70 substrate binding. Mol. Cell. Biol. 24, 3928–3937 (2004).
Tuite, M. F., Mundy, C. R. & Cox, B. S. Agents that cause a high frequency of genetic change from [psi+] to [psi−] in Saccharomyces cerevisiae. Genetics 98, 691–711 (1981).
Ferreira, P. C., Ness, F., Edwards, S. R., Cox, B. S. & Tuite, M. F. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol. Microbiol. 40, 1357–1369 (2001).
Jung, G., Jones, G. & Masison, D. C. Amino acid residue 184 of yeast Hsp104 chaperone is critical for prion-curing by guanidine, prion propagation, and thermotolerance. Proc. Natl Acad. Sci. USA 99, 9936–9941 (2002). This report demonstrated that uanidine cures prions by inhibiting Hsp104.
Glover, J. R. & Lindquist, S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94, 73–82 (1998). Reported that chaperones interact in disaggregating proteins.
Paushkin, S. V., Kushnirov, V. V., Smirnov, V. N. & Ter-Avanesyan, M. D. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 15, 3127–3134 (1996).
Ness, F., Ferreira, P., Cox, B. S. & Tuite, M. F. Guanidine hydrochloride inhibits the generation of prion 'seeds' but not prion protein aggregation in yeast. Mol. Cell. Biol. 22, 5593–5605 (2002).
Cox, B. S., Ness, F. & Tuite, M. F. Analysis of the generation and segregation of propagons: entities that propagate the [PSI+] prion in yeast. Genetics 165, 23–33 (2003).
Tuite, M. F. & Koloteva-Levin, N. Propagating prions in fungi and mammals. Mol. Cell 14, 541–552 (2004).
Schwimmer, C. & Masison, D. C. Antagonistic interactions between yeast [PSI+] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol. Cell. Biol. 22, 3590–3598 (2002).
Hall, D. & Edskes, H. K. Silent prions lying in wait: a two-hit model of prion/amyloid formation and infection. J. Mol. Biol. 336, 775–786 (2004).
Collins, S. R., Douglass, A., Vale, R. D. & Weissman, J. S. Mechanism of prion propagation: amyloid growth occurs by monomer addition. PLoS Biol. 2, 1582–1590 (2004).
Serio, T. R. et al. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science 289, 1317–1321 (2000).
Bruce, M. E., McConnell, I., Fraser, H. & Dickinson, A. G. The disease characteristics of different strains of scrapie in Sinc congenic mouse lines: implications for the nature of the agent and host control of pathogenesis. J. Gen. Virol. 72, 595–603 (1991).
Derkatch, I. L., Chernoff, Y. O., Kushnirov, V. V., Inge-Vechtomov, S. G. & Liebman, S. W. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 144, 1375–1386 (1996). The original description of the [ PIN+] prion.
Bradley, M. E., Edskes, H. K., Hong, J. Y., Wickner, R. B. & Liebman, S. W. Interactions among prions and prion 'strains' in yeast. Proc. Natl Acad. Sci. USA 99, 16392–16399 (2002).
King, C. Y. Supporting the structural basis of prion strains: induction and identification of [PSI] variants. J. Mol. Biol. 307, 1247–1260 (2001).
Pattison, I. H. in Slow, Latent and Temperate Virus Infection (eds Gajdusek, D. C., Gibbs, C. J. & Alpers, M. P.) 249–257 (US Government Printing Office, Washington DC, 1965).
Collinge, J. Variant Creutzfeldt-Jakob disease. Lancet 354, 317–323 (1999).
Kushnirov, V. V., Kochneva-Pervukhova, N. V., Cechenova, M. B., Frolova, N. S. & Ter-Avanesyan, M. D. Prion properties of the Sup35 protein of yeast Pichia methanolica. EMBO J. 19, 324–331 (2000).
Chernoff, Y. O. et al. Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Molec. Microbiol. 35, 865–876 (2000).
Santoso, A., Chien, P., Osherovich, L. Z. & Weissman, J. S. Molecular basis of a yeast prion species barrier. Cell 100, 277–288 (2000).
Nakayashiki, T., Ebihara, K., Bannai, H. & Nakamura, Y. Yeast [PSI+] 'prions' that are crosstransmissible and susceptible beyond a species barrier through a quasi-prion state. Mol. Cell 7, 1121–1130 (2001).
Tanaka, M., Chien, P., Yonekura, K. & Weissman, J. S. Mechanism of cross-species prion transmission: an infectious conformation compatible with two highly divergent yeast prion proteins. Cell 121, 49–62 (2005).
Osherovich, L. Z. & Weissman, J. S. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI+] prion. Cell 106, 183–194 (2001).
Vitrenko, Y. A., Gracheva, E. O., Richmond, J. E. & Leibman, S. W. Visualization of aggregation of the Rnq1 prion domain and cross-seeding interactions with Sup35NM. J. Biol. Chem. 282, 1779–1787 (2007).
Bradley, M. E. & Liebman, S. W. Destabilizing interactions among [PSI+] and [PIN+] yeast prion variants. Genetics 165, 1675–1685 (2003).
Chernoff, Y. O., Newnam, G. P., Kumar, J., Allen, K. & Zink, A. D. Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone Ssb in formation, stability and toxicity of the [PSI+] prion. Mol. Cell. Biol. 19, 8103–8112 (1999).
Allen, K. D., Chernova, T. A., Tennant, E. P., Wilkinson, K. D. & Chernoff, Y. O. Effects of ubiquitin system alterations on the formation and loss of a yeast prion. J. Biol. Chem. 282, 3004–3013 (2006).
Ganusova, E. E. et al. Modulation of prion formation, aggregation, and toxicity by the actin cytoskeleton in yeast. Mol. Cell. Biol. 26, 617–629 (2006).
Chapman, M. R. et al. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295, 851–855 (2002).
Podrabsky, J. E., Carpenter, J. F. & Hand, S. C. Survival of water stress in annual fish embryos: dehydration avoidance and egg amyloid fibers. Am. J. Physiol. Regul. Integr. Comp. Physiol. 280, R123–R131 (2001).
Wosten, H. A. & de Vocht, M. L. Hydrophobins, the fungal coat unravelled. Biochim. Biophys. Acta 1469, 79–86 (2000).
Berson, J. F. et al. Proprotein convertase cleavage liberates a fibrillogenic fragment of a resident glycoprotein to initiate melanosome biogenesis. J. Cell Biol. 161, 521–533 (2003).
Wickner, R. B. A new prion controls fungal cell fusion incompatibility. Proc. Natl Acad. Sci. USA 94, 10012–10014 (1997).
True, H. L. & Lindquist, S. L. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407, 477–483 (2000).
Partridge, L. & Barton, N. H. Evolving evolvability. Nature 407, 457–458 (2000).
Nakayashiki, T., Kurtzman, C. P., Edskes, H. K. & Wickner, R. B. Yeast prions [URE3] and [PSI+] are diseases. Proc. Natl Acad. Sci. USA 102, 10575–10580 (2005). Showed that [URE3] and [ PSI+] are disease agents.
Resende, C. G., Outeiro, T. F., Sands, L., Lindquist, S. & Tuite, M. F. Prion protein gene polymorphisms in Saccharomyces cerevisiae. Mol. Microbiol. 49, 1005–1017 (2003).
Dalstra, H. J. P., Swart, K., Debets, A. J. M., Saupe, S. J. & Hoekstra, R. F. Sexual transmission of the [Het-s] prion leads to meiotic drive in Podospora anserina. Proc. Natl Acad. Sci. USA 100, 6616–6621 (2003).
Edskes, H. K. & Wickner, R. B. Conservation of a portion of the Saccharomyces cerevisiae Ure2p prion domain that interacts with the full- length protein. Proc. Natl Acad. Sci. USA 99 (Suppl. 4), 16384–16391 (2002).
Talarek, N., Maillet, L., Cullin, C. & Aigle, M. The [URE3] prion is not conserved among Saccharomyces species. Genetics 171, 23–54 (2005).
Shewmaker, F., Mull, L., Nakayashiki, T., Masison, D. C. & Wickner, R. B. Ure2p function is enhanced by its prion domain in Saccharomyces cerevisiae. Genetics 16 May 2007 (doi:10.1534 genetics.107.074153).
Gagny, B. & Silar, P. Identification of the genes encoding the cytosolic translation release factors from Podospora anserina and analysis of their role during the life cycle. Genetics 149, 1763–1775 (1988).
Urakov, V. N. et al. N-terminal region of Saccharomyces cerevisiae eRF3 is essential for the functioning of the eRF1/eRF3 complex beyond translation termination. BMC Mol. Biol. 7, 34–46 (2006).
Bach, S. et al. Isolation of drugs active against mammalian prions using a yeast-based screening assay. Nature Biotechnol. 21, 1075–1081 (2003). This report used yeast to find drugs against prion diseases of mammals.
Creutzfeldt, H. G. Uber eine eigenartige herdformige Erkrankung des Zentralnervensystems. Neurol. Psychiat. 57, 1–18 (1920).
Jakob, A. Uber eigenartige Erkrankung des Zentalnervensystems mit bemerkenswertem anatomischen Befunde (Spastische Pseudosklarose-encephalomyopathie mit disseminierten Degenerationsherden). Neurol. Psychiatr. 64, 147–228 (1921).
Cuille, J. & Chelle, P. L. Pathologie animale. La maladie dite tremblant du mouton est-elle inoculable? Compt. Rend. Acad. Sci. (Paris) 203, 1552–1554 (1936).
Cuille, J. & Chelle, P. L. Experimental transmission of trembling to the goat. C. R. Seances Acad. Sci. 208, 1058–1060 (1939).
Rizet, G. Les phenomenes de barrage chez Podospora anserina: analyse genetique des barrages entre les souches s et S. Rev. Cytol. Biol. Veg. 13, 51–92 (1952).
Zigas, V. & Gajdusek, D. C. Kuru: clinical study of a new syndrome resembling paralysis agitans in natives of the Eastern Highlands of Australian New Guinea. Med. J. Aust. 2, 745–754 (1957).
Hadlow, W. J. Scrapie and kuru. Lancet 2, 289–290 (1959).
Chandler, R. L. Encephalopathy in mice produced by inoculation with scrapie brain material. Lancet 1, 107–108 (1961).
Cox, B. S. PSI, a cytoplasmic suppressor of super-suppressor in yeast. Heredity 20, 505–521 (1965).
Gajdusek, D. C., Gibbs, C. J. & Alpers, M. Experimental transmission of a kuru-like syndrome to chimpanzees. Nature 209, 794–796 (1966).
Alper, T., Haig, D. A. & Clarke, M. C. The exceptionally small size of the scrapie agent. Biochem. Biophys. Res. Commun. 22, 278–284 (1966).
Griffith, J. S. Self-replication and scrapie. Nature 215, 1043–1044 (1967).
Dickinson, A. G., Meikle, V. M. H. & Fraser, H. Identification of a gene which controls the incubation period of some strains of scrapie in mice. J. Comp. Path. 78, 293–299 (1968).
Lacroute, F. Non-mendelian mutation allowing ureidosuccinic acid uptake in yeast. J. Bacteriol. 106, 519–522 (1971).
Bolton, D. C., McKinley, M. P. & Prusiner, S. B. Identification of a protein that purifies with the scrapie prion. Science 218, 1309–1311 (1982).
Oesch, B. et al. A cellular gene encodes scrapie PrP 27–30 protein. Cell 40, 735–746 (1985).
Chesebro, B. et al. Identification of scrapie prion protein-specific mRNA in scrapie-infected brain. Nature 315, 331–333 (1985).
Carlson, G. A. et al. Linkagae of prion protein and scrapie incubation time genes. Cell 46, 503–511 (1986).
Wells, G. A. H. et al. A novel progressive spongiform encephalopathy in cattle. Vet. Rec. 121, 419–420 (1987).
Owen, F. et al. Insertion in prion protein gene in familial Creutzfeldt-Jakob disease. Lancet 1, 51–52 (1989).
Hsiao, K. et al. Linkage of a prion protein missense variant to Gerstmann-Straussler syndrome. Nature 338, 342–345 (1989).
Bueler, H. et al. Mice devoid of PrP are resistant to scrapie. Cell 73, 1339–1347 (1993).
Bueler, H. et al. Normal development and behavior of mice lacking the neuronal cell-surface PrP protein. Nature 356, 577–582 (1992).
Britton, T. C., AlSarraj, S., Shaw, C., Campbell, T. & Collinge, J. Sporadic Creutzfeldt-Jakob disease in a 16-year-old in the UK. Lancet 346, 1155 (1995).
Acknowledgements
This research was supported by the Intramural Research Program of the National Institutes of Health, The National Institute of Diabetes and Digestive and Kidney Diseases.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
DATABASES
Entrez Genome Project
Entrez Protein
Glossary
- Prion
-
An infectious protein that does not require a nucleic acid for infectivity.
- Amyloid
-
A filamentous form of protein with a cross β-sheet structure, meaning that the β-strands are perpendicular to the long axis of the filaments.
- Non-chromosomal (cytoplasmic) genetic element
-
A gene or replicon that is inherited or transmitted independently of the chromosomes, such as the mitochondrial genome, the 2 μm plasmid, a yeast virus or a prion.
- Gene gun
-
A device that uses a pneumatic gun to propel gold particles coated with DNA or protein into cells to genetically transform them.
- Prion seed
-
An amyloid fragment that can grow, become fragmented again and thus propagate the prion. Similarly, active enzyme molecules of the [β] and [C] prions can act as seeds.
- Parallel in-register β-sheet
-
A β-sheet in which each residue is aligned with the same residue of the adjacent strand.
- Nuclear magnetic resonance
-
(NMR). A technique in which the distances between labelled nuclei can be measured by the rate of decay of the magnetic signal due to dipole–dipole coupling.
Rights and permissions
About this article
Cite this article
Wickner, R., Edskes, H., Shewmaker, F. et al. Prions of fungi: inherited structures and biological roles. Nat Rev Microbiol 5, 611–618 (2007). https://doi.org/10.1038/nrmicro1708
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1708
This article is cited by
-
Divergent CPEB prion-like domains reveal different assembly mechanisms for a generic amyloid-like fold
BMC Biology (2021)
-
MED15 prion-like domain forms a coiled-coil responsible for its amyloid conversion and propagation
Communications Biology (2021)
-
Innate immunity to prions: anti-prion systems turn a tsunami of prions into a slow drip
Current Genetics (2021)
-
RNA modulates aggregation of the recombinant mammalian prion protein by direct interaction
Scientific Reports (2019)
-
Prions are affected by evolution at two levels
Cellular and Molecular Life Sciences (2016)