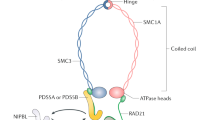
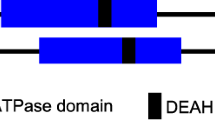
Abstract
Cohesin is a conserved multisubunit protein complex with diverse cellular roles, making key contributions to the coordination of chromosome segregation, the DNA damage response and chromatin regulation by epigenetic mechanisms. Much has been learned in recent years about the roles of cohesin in a physiological context, whereas its potential and emerging role in tumour initiation and/or progression has received relatively little attention. In this Opinion article we examine how cohesin deregulation could contribute to cancer development on the basis of its physiological roles.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
26 February 2011
In figure 4 the REC8 locus was labelled 12q11.2 intsead of 14q11.2. This has been corrected on both the html and pdf versions.
References
Michaelis, C., Ciosk, R. & Nasmyth, K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91, 35–45 (1997).
Guacci, V., Koshland, D. & Strunnikov, A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91, 47–57 (1997).
Sonoda, E. et al. Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev. Cell 1, 759–770 (2001).
Hauf, S., Waizenegger, I. C. & Peters, J. M. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science 293, 1320–1323 (2001).
Watrin, E. & Peters, J. M. Cohesin and DNA damage repair. Exp. Cell Res. 312, 2687–2693 (2006).
Covo, S., Westmoreland, J. W., Gordenin, D. A. & Resnick, M. A. Cohesin Is limiting for the suppression of DNA damage-induced recombination between homologous chromosomes. PLoS Genet. 6, e1001006 (2010).
Jessberger, R. Cohesin's dual role in the DNA damage response: repair and checkpoint activation. EMBO J. 28, 2491–2493 (2009).
Bauerschmidt, C. et al. Cohesin promotes the repair of ionizing radiation-induced DNA double-strand breaks in replicated chromatin. Nucleic Acids Res. 38, 477–487 (2010).
Jessberger, R., Podust, V., Hubscher, U. & Berg, P. A mammalian protein complex that repairs double-strand breaks and deletions by recombination. J. Biol. Chem. 268, 15070–15079 (1993).
Kitagawa, R., Bakkenist, C. J., McKinnon, P. J. & Kastan, M. B. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes Dev. 18, 1423–1438 (2004).
Kim, S. T., Xu, B. & Kastan, M. B. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 16, 560–570 (2002).
Xu, H. et al. Rad21-cohesin haploinsufficiency impedes DNA repair and enhances gastrointestinal radiosensitivity in mice. PLoS One 5, e12112 (2010).
Horsfield, J. A. et al. Cohesin-dependent regulation of Runx genes. Development 134, 2639–2649 (2007).
Bausch, C. et al. Transcription alters chromosomal locations of cohesin in Saccharomyces cerevisiae. Mol. Cell Biol. 27, 8522–8532 (2007).
Dorsett, D. Cohesin, gene expression and development: lessons from Drosophila. Chromosome Res. 17, 185–200 (2009).
Liu, J. et al. Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol. 7, e1000119 (2009).
Birkenbihl, R. P. & Subramani, S. Cloning and characterization of rad21 an essential gene of Schizosaccharomyces pombe involved in DNA double-strand-break repair. Nucleic Acids Res. 20, 6605–6611 (1992).
Atienza, J. M. et al. Suppression of RAD21 gene expression decreases cell growth and enhances cytotoxicity of etoposide and bleomycin in human breast cancer cells. Mol. Cancer Ther. 4, 361–368 (2005).
Xu, H. et al. Enhanced RAD21 cohesin expression confers poor prognosis and resistance to chemotherapy in high grade luminal, basal and HER2 breast cancers. Breast Cancer Resarch 21 Jan 2011 (doi: 10.1186/bcr2814).
Ghiselli, G. & Iozzo, R. V. Overexpression of bamacan/SMC3 causes transformation. J. Biol. Chem. 275, 20235–20238 (2000).
Rhodes, D. R. et al. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc. Natl Acad. Sci. USA 101, 9309–9314 (2004).
Yu, R., Lu, W., Chen, J., McCabe, C. J. & Melmed, S. Overexpressed pituitary tumor-transforming gene causes aneuploidy in live human cells. Endocrinology 144, 4991–4998 (2003).
Zhang, N. et al. Overexpression of Separase induces aneuploidy and mammary tumorigenesis. Proc. Natl Acad. Sci. USA 105, 13033–13038 (2008).
Salehi, F., Kovacs, K., Scheithauer, B. W., Lloyd, R. V. & Cusimano, M. Pituitary tumor-transforming gene in endocrine and other neoplasms: a review and update. Endocr. Relat Cancer 15, 721–743 (2008).
Oikawa, K. et al. Expression of a novel human gene, human wings apart-like (hWAPL), is associated with cervical carcinogenesis and tumor progression. Cancer Res. 64, 3545–3549 (2004).
van 't Veer, L. J. et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415, 530–536 (2002).
Yamamoto, G. et al. Correlation of invasion and metastasis of cancer cells, and expression of the RAD21 gene in oral squamous cell carcinoma. Virchows Arch. 448, 435–441 (2006).
Porkka, K. P., Tammela, T. L., Vessella, R. L. & Visakorpi, T. RAD21 and KIAA0196 at 8q24 are amplified and overexpressed in prostate cancer. Genes Chromosom. Cancer 39, 1–10 (2004).
Roe, O. D. et al. Genome-wide profile of pleural mesothelioma versus parietal and visceral pleura: the emerging gene portrait of the mesothelioma phenotype. PLoS ONE 4, e6554 (2009).
Ryu, B., Kim, D. S., Deluca, A. M. & Alani, R. M. Comprehensive expression profiling of tumor cell lines identifies molecular signatures of melanoma progression. PLoS ONE 2, e594 (2007).
Iwaizumi, M. et al. Human Sgo1 Down-regulation Leads to Chromosomal Instability in Colorectal Cancer. Gut 58, 249–260 (2008).
Hagemann, C. et al. The cohesin-interacting protein, precocious dissociation of sisters 5A/sister chromatid cohesion protein 112, is up-regulated in human astrocytic tumors. Int. J. Mol. Med. 27, 39–51 (2011).
Barber, T. D. et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc. Natl Acad. Sci. USA 105, 3443–3448 (2008).
Nasmyth, K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35, 673–745 (2001).
Watrin, E. & Peters, J. M. The cohesin complex is required for the DNA damage-induced G2/M checkpoint in mammalian cells. EMBO J. 28, 2625–2635 (2009).
Hakimi, M. A. et al. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature 418, 994–998 (2002).
Glynn, E. F. et al. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2, E259 (2004).
Lengronne, A. et al. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430, 573–578 (2004).
Kang, H. & Lieberman, P. M. Cell cycle control of Kaposi's sarcoma-associated herpesvirus latency transcription by CTCF-cohesin interactions. J. Virol. 83, 6199–6210 (2009).
Bowers, S. R. et al. A conserved insulator that recruits CTCF and cohesin exists between the closely related but divergently regulated interleukin-3 and granulocyte-macrophage colony-stimulating factor genes. Mol. Cell Biol. 29, 1682–1693 (2009).
Wendt, K. S. et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451, 796–801 (2008).
Stedman, W. et al. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 27, 654–666 (2008).
Kagey, M. H. et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 (2010).
Rhodes, J. M. et al. Positive regulation of c-Myc by cohesin is direct, and evolutionarily conserved. Dev. Biol. 344, 637–649 (2010).
Wendt, K. S. & Peters, J. M. How cohesin and CTCF cooperate in regulating gene expression. Chromosome Res. 17, 201–214 (2009).
Dorsett, D. et al. Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development 132, 4743–4753 (2005).
Nasmyth, K. & Haering, C. H. Cohesin: Its Roles and Mechanisms. Annu. Rev. Genet. 43, 525–558 (2009).
Peters, J. M., Tedeschi, A. & Schmitz, J. The cohesin complex and its roles in chromosome biology. Genes Dev. 22, 3089–3114 (2008).
Wood, A. J., Severson, A. F. & Meyer, B. J. Condensin and cohesin complexity: the expanding repertoire of functions. Nature Rev. Genet. 11, 391–404 (2010).
McKay, M. J. et al. Sequence conservation of the rad21 Schizosaccharomyces pombe DNA double-strand break repair gene in human and mouse. Genomics 36, 305–315 (1996).
Jessberger, R., Riwar, B., Baechtold, H. & Akhmedov, A. T. SMC proteins constitute two subunits of the mammalian recombination complex RC-1. EMBO J. 15, 4061–4068 (1996).
Parisi, S. et al. Rec8p, a meiotic recombination and sister chromatid cohesion phosphoprotein of the Rad21p family conserved from fission yeast to humans. Mol. Cell Biol. 19, 3515–3528 (1999).
Hirano, T. The ABCs of SMC proteins: two-armed ATPases for chromosome condensation, cohesion, and repair. Genes Dev. 16, 399–414 (2002).
Jessberger, R. The many functions of SMC proteins in chromosome dynamics. Nature Rev. Mol. Cell Biol. 3, 767–778 (2002).
Hirano, T., Mitchison, T. J. & Swedlow, J. R. The SMC family: from chromosome condensation to dosage compensation. Curr. Opin. Cell Biol. 7, 329–336 (1995).
Hirano, T. At. the heart of the chromosome: SMC proteins in action. Nature Rev. Mol. Cell Biol. 7, 311–322 (2006).
Haering, C. H., Lowe, J., Hochwagen, A. & Nasmyth, K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9, 773–788 (2002).
Haering, C. H. et al. Structure and stability of cohesin's Smc1-kleisin interaction. Mol. Cell 15, 951–964 (2004).
Haering, C. H., Farcas, A. M., Arumugam, P., Metson, J. & Nasmyth, K. The cohesin ring concatenates sister DNA molecules. Nature 454, 297–301 (2008).
Zhang, N. et al. A handcuff model for the cohesin complex. J. Cell Biol. 183, 1019–1031 (2008).
Huang, C. E., Milutinovich, M. & Koshland, D. Rings, bracelet or snaps: fashionable alternatives for Smc complexes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360, 537–542 (2005).
Watanabe, Y. & Nurse, P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature 400, 461–464 (1999).
Molnar, M., Bahler, J., Sipiczki, M. & Kohli, J. The rec8 gene of Schizosaccharomyces pombe is involved in linear element formation, chromosome pairing and sister-chromatid cohesion during meiosis. Genetics 141, 61–73 (1995).
Tomonaga, T. et al. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 14, 2757–2770 (2000).
Pasierbek, P. et al. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 15, 1349–1360 (2001).
Revenkova, E., Eijpe, M., Heyting, C., Gross, B. & Jessberger, R. Novel meiosis-specific isoform of mammalian SMC1. Mol. Cell Biol. 21, 6984–6998 (2001).
Canudas, S. & Smith, S. Differential regulation of telomere and centromere cohesion by the Scc3 homologues SA1 and SA2, respectively, in human cells. J. Cell Biol. 187, 165–173 (2009).
Pezzi, N. et al. STAG3, a novel gene encoding a protein involved in meiotic chromosome pairing and location of STAG3-related genes flanking the Williams-Beuren syndrome deletion. FASEB J. 14, 581–592 (2000).
Parra, M. T. et al. Involvement of the cohesin Rad21 and SCP3 in monopolar attachment of sister kinetochores during mouse meiosis I. J. Cell Sci. 117, 1221–1234 (2004).
Xu, H. et al. A new role for the mitotic RAD21/SCC1 cohesin in meiotic chromosome cohesion and segregation in the mouse. EMBO Rep. 5, 378–384 (2004).
Kueng, S. et al. Wapl controls the dynamic association of cohesin with chromatin. Cell 127, 955–967 (2006).
Ciosk, R. et al. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell 5, 243–254 (2000).
Rollins, R. A., Korom, M., Aulner, N., Martens, A. & Dorsett, D. Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol. Cell Biol. 24, 3100–3111 (2004).
Ivanov, D. et al. Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion. Curr. Biol. 12, 323–328 (2002).
Hou, F. & Zou, H. Two human orthologues of Eco1/Ctf7 acetyltransferases are both required for proper sister-chromatid cohesion. Mol. Biol. Cell 16, 3908–3918 (2005).
Tanaka, K., Hao, Z., Kai, M. & Okayama, H. Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. EMBO J. 20, 5779–5790 (2001).
Hartman, T., Stead, K., Koshland, D. & Guacci, V. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J. Cell Biol. 151, 613–626 (2000).
Panizza, S., Tanaka, T., Hochwagen, A., Eisenhaber, F. & Nasmyth, K. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr. Biol. 10, 1557–1564 (2000).
Gandhi, R., Gillespie, P. J. & Hirano, T. Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr. Biol. 16, 2406–2417 (2006).
Terret, M. E., Sherwood, R., Rahman, S., Qin, J. & Jallepalli, P. V. Cohesin acetylation speeds the replication fork. Nature 462, 231–234 (2009).
Peters, J. M. & Bhaskara, V. Cohesin acetylation: from antiestablishment to establishment. Mol. Cell 34, 1–2 (2009).
Zhang, J. et al. Acetylation of Smc3 by Eco1 is required for S. phase sister chromatid cohesion in both human and yeast. Mol. Cell 31, 143–151 (2008).
Wang, Z., Castano, I. B., De Las Penas, A., Adams, C. & Christman, M. F. Pol κ: A DNA polymerase required for sister chromatid cohesion. Science 289, 774–779 (2000).
Hanna, J. S., Kroll, E. S., Lundblad, V. & Spencer, F. A. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol. Cell Biol. 21, 3144–3158 (2001).
Mayer, M. L., Gygi, S. P., Aebersold, R. & Hieter, P. Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol. Cell 7, 959–970 (2001).
Uhlmann, F., Lottspeich, F. & Nasmyth, K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400, 37–42 (1999).
Rao, H., Uhlmann, F., Nasmyth, K. & Varshavsky, A. Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature 410, 955–959 (2001).
Ciosk, R. et al. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell 93, 1067–1076 (1998).
Sumara, I., Vorlaufer, E., Gieffers, C., Peters, B. H. & Peters, J. M. Characterization of vertebrate cohesin complexes and their regulation in prophase. J. Cell Biol. 151, 749–762 (2000).
Waizenegger, I. C., Hauf, S., Meinke, A. & Peters, J. M. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell 103, 399–410 (2000).
Hauf, S. et al. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 3, e69 (2005).
McGuinness, B. E., Hirota, T., Kudo, N. R., Peters, J. M. & Nasmyth, K. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 3, e86 (2005).
Kitajima, T. S., Hauf, S., Ohsugi, M., Yamamoto, T. & Watanabe, Y. Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting Shugoshin localization. Curr. Biol. 15, 353–359 (2005).
Pouwels, J. et al. Shugoshin 1 plays a central role in kinetochore assembly and is required for kinetochore targeting of Plk1. Cell Cycle 6, 1579–1585 (2007).
Nakajima, M. et al. The complete removal of cohesin from chromosome arms depends on separase. J. Cell Sci. 120, 4188–4196 (2007).
Rivera, T. & Losada, A. Shugoshin and PP2A, shared duties at the centromere. Bioessays 28, 775–779 (2006).
Dai, J., Sullivan, B. A. & Higgins, J. M. Regulation of mitotic chromosome cohesion by Haspin and Aurora, B. Dev. Cell 11, 741–750 (2006).
Takata, H. et al. PHB2 protects sister-chromatid cohesion in mitosis. Curr. Biol. 17, 1356–1361 (2007).
Zou, H., McGarry, T. J., Bernal, T. & Kirschner, M. W. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science 285, 418–422 (1999).
Stemmann, O., Zou, H., Gerber, S. A., Gygi, S. P. & Kirschner, M. W. Dual inhibition of sister chromatid separation at metaphase. Cell 107, 715–726 (2001).
Boos, D., Kuffer, C., Lenobel, R., Korner, R. & Stemmann, O. Phosphorylation-dependent binding of cyclin B1 to a Cdc6-like domain of human separase. J. Biol. Chem. 283, 816–823 (2007).
Fang, G., Yu, H. & Kirschner, M. W. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 12, 1871–1883 (1998).
Shah, J. V. & Cleveland, D. W. Waiting for anaphase: Mad2 and the spindle assembly checkpoint. Cell 103, 997–1000 (2000).
Michel, L. S. et al. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature 409, 355–359 (2001).
Wang, Z., Yu, R. & Melmed, S. Mice lacking pituitary tumor transforming gene show testicular and splenic hypoplasia, thymic hyperplasia, thrombocytopenia, aberrant cell cycle progression, and premature centromere division. Mol. Endocrinol. 15, 1870–1879 (2001).
Musio, A. et al. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nature Genet. 38, 528–530 (2006).
Deardorff, M. A. et al. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of cornelia de Lange syndrome with predominant mental retardation. Am. J. Hum. Genet. 80, 485–494 (2007).
Pie, J. et al. Mutations and variants in the cohesion factor genes NIPBL, SMC1A, and SMC3 in a cohort of 30 unrelated patients with Cornelia de Lange syndrome. Am. J. Med. Genet. A 152A, 924–929 (2010).
Vega, H. et al. Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nature Genet. 37, 468–470 (2005).
Vega, H. et al. Phenotypic variability in 49 cases of ESCO2 mutations, including novel missense and codon deletion in the acetyltransferase domain, correlates with ESCO2 expression and establishes the clinical criteria for Roberts syndrome. J. Med. Genet. 47, 30–37 (2010).
Krantz, I. D. et al. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nature Genet. 36, 631–635 (2004).
van der Lelij, P. et al. Warsaw breakage syndrome, a cohesinopathy associated with mutations in the XPD helicase family member DDX11/ChlR1. Am. J. Hum. Genet. 86, 262–266 (2010).
Newman, J. J. & Young, R. A. Connecting Transcriptional Control to Chromosome Structure and Human Disease. Cold Spring Harb. Symp. Quant. Biol. 5 Jan 2011 (doi:10.1101/sqb.2010.75.016).
Mannini, L., Menga, S. & Musio, A. The expanding universe of cohesin functions: a new genome stability caretaker involved in human disease and cancer. Hum. Mutat. 31, 623–630 (2010).
Xu, H., Beasley, M. D., Warren, W. D., van der Horst, G. T. & McKay, M. J. Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev. Cell 8, 949–961 (2005).
Zhang, B. et al. Mice lacking sister chromatid cohesion protein PDS5B exhibit developmental abnormalities reminiscent of Cornelia de Lange syndrome. Development 134, 3191–3201 (2007).
Kawauchi, S. et al. Multiple organ system defects and transcriptional dysregulation in the Nipbl(+/−) mouse, a model of Cornelia de Lange Syndrome. PLoS Genet. 5, e1000650 (2009).
Parry, D. M., Mulvihill, J. J., Tsai, S. E., Kaiser-Kupfer, M. I. & Cowan, J. M. SC phocomelia syndrome, premature centromere separation, and congenital cranial nerve paralysis in two sisters, one with malignant melanoma. Am. J. Med. Genet. 24, 653–672 (1986).
Wenger, S. L. et al. Rhabdomyosarcoma in Roberts syndrome. Cancer Genet. Cytogenet. 31, 285–289 (1988).
Turnbull, C. & Rahman, N. Genetic predisposition to breast cancer: past, present, and future. Annu. Rev. Genomics Hum. Genet. 9, 321–345 (2008).
Van den Berg, D. J. & Francke, U. Sensitivity of Roberts syndrome cells to gamma radiation, mitomycin C, and protein synthesis inhibitors. Somat. Cell. Mol. Genet. 19, 377–392 (1993).
Goh, E. S. et al. The Roberts syndrome/SC phocomelia spectrum--a case report of an adult with review of the literature. Am. J. Med. Genet. A 152A, 472–478 (2010).
Liu, J. et al. SMC1A expression and mechanism of pathogenicity in probands with X-linked Cornelia de Lange Syndrome. Hum. Mutat. 30, 1535–1542 (2009).
Revenkova, E. et al. Cornelia de Lange syndrome mutations in SMC1A or SMC3 affect binding to, D. N. A. Hum. Mol. Genet. 18, 418–427 (2009).
Gerkes, E. H., van der Kevie-Kersemaekers, A. M., Yakin, M., Smeets, D. F. & van Ravenswaaij-Arts, C. M. The importance of chromosome studies in Roberts syndrome/SC phocomelia and other cohesinopathies. Eur. J. Med. Genet. 53, 40–44 (2009).
Tomkins, D. J. Premature centromere separation and the prenatal diagnosis of Roberts syndrome. Prenat. Diagn 9, 450–452 (1989).
Burns, M. A. & Tomkins, D. J. Hypersensitivity to mitomycin C cell-killing in Roberts syndrome fibroblasts with, but not without, the heterochromatin abnormality. Mutat. Res. 216, 243–249 (1989).
van der Lelij, P. et al. The cellular phenotype of Roberts syndrome fibroblasts as revealed by ectopic expression of ESCO2. PLoS ONE 4, e6936 (2009).
Kajii, T. et al. Cancer-prone syndrome of mosaic variegated aneuploidy and total premature chromatid separation: report of five infants. Am. J. Med. Genet. 104, 57–64 (2001).
Callier, P. et al. Microcephaly is not mandatory for the diagnosis of mosaic variegated aneuploidy syndrome. Am. J. Med. Genet. A 137, 204–207 (2005).
Mannini, L., Liu, J., Krantz, I. D. & Musio, A. Spectrum and consequences of SMC1A mutations: the unexpected involvement of a core component of cohesin in human disease. Hum. Mutat. 31, 5–10 (2009).
Hallson, G. et al. The Drosophila cohesin subunit Rad21 is a trithorax group (trxG) protein. Proc. Natl Acad. Sci. USA 105, 12405–12410 (2008).
Tfelt-Hansen, J., Kanuparthi, D. & Chattopadhyay, N. The emerging role of pituitary tumor transforming gene in tumorigenesis. Clin. Med. Res. 4, 130–137 (2006).
Kakar, S. S. & Malik, M. T. Suppression of lung cancer with siRNA targeting PTTG. Int. J. Oncol. 29, 387–395 (2006).
El-Naggar, S. M., Malik, M. T. & Kakar, S. S. Small interfering RNA against PTTG: a novel therapy for ovarian cancer. Int. J. Oncol. 31, 137–143 (2007).
Ramaswamy, S., Ross, K. N., Lander, E. S. & Golub, T. R. A molecular signature of metastasis in primary solid tumors. Nature Genet. 33, 49–54 (2003).
Meyer, R. et al. Overexpression and mislocalization of the chromosomal segregation protein separase in multiple human cancers. Clin. Cancer Res. 15, 2703–2710 (2009).
Pino, M. S. & Chung, D. C. The chromosomal instability pathway in colon cancer. Gastroenterology 138, 2059–2072 (2010).
Hoque, M. T. & Ishikawa, F. Human chromatid cohesin component hRad21 is phosphorylated in M phase and associated with metaphase centromeres. J. Biol. Chem. 276, 5059–5067 (2001).
Chen, F. et al. Caspase proteolysis of the cohesin component RAD21 promotes apoptosis. J. Biol. Chem. 277, 16775–16781 (2002).
Pati, D., Zhang, N. & Plon, S. E. Linking sister chromatid cohesion and apoptosis: role of Rad21. Mol. Cell Biol. 22, 8267–8277 (2002).
Heo, S. J., Tatebayashi, K., Kato, J. & Ikeda, H. The RHC21 gene of budding yeast, a homologue of the fission yeast rad21+ gene, is essential for chromosome segregation. Mol. Gen. Genet. 257, 149–156 (1998).
Jallepalli, P. V. et al. Securin is required for chromosomal stability in human cells. Cell 105, 445–457 (2001).
Waizenegger, I., Gimenez-Abian, J. F., Wernic, D. & Peters, J. M. Regulation of human separase by securin binding and autocleavage. Curr. Biol. 12, 1368–1378 (2002).
Begus-Nahrmann, Y. et al. p53 deletion impairs clearance of chromosomal-instable stem cells in aging telomere-dysfunctional mice. Nature Genet. 41, 1138–1143 (2009).
Diaz-Martinez, L. A. & Clarke, D. J. Chromosome cohesion and the spindle checkpoint. Cell Cycle 8, 2733–2740 (2009).
Holt, J. E. & Jones, K. T. Control of homologous chromosome division in the mammalian oocyte. Mol. Hum. Reprod. 15, 139–147 (2009).
Yu, H. & Tang, Z. Bub1 multitasking in mitosis. Cell Cycle 4, 262–265 (2005).
Soshnikova, N. & Duboule, D. Epigenetic regulation of Hox gene activation: the waltz of methyls. Bioessays 30, 199–202 (2008).
Martin-Perez, D., Piris, M. A. & Sanchez-Beato, M. Polycomb proteins in hematologic malignancies. Blood 116, 5465–5475 (2010).
Nikolaev, L. G., Akopov, S. B., Didych, D. A. & Sverdlov, E. D. Vertebrate protein CTCF and its multiple roles in a large-scale regulation of genome activity. Curr. Genomics 10, 294–302 (2009).
Filippova, G. N. Genetics and epigenetics of the multifunctional protein CTCF. Curr. Top. Dev. Biol. 80, 337–360 (2008).
Hadjur, S. et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460, 410–413 (2009).
Nativio, R. et al. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 5, e1000739 (2009).
Cui, H. Loss of imprinting of IGF2 as an epigenetic marker for the risk of human cancer. Dis. Markers 23, 105–112 (2007).
Jelinic, P. & Shaw, P. Loss of imprinting and cancer. J. Pathol. 211, 261–268 (2007).
Miele, A. & Dekker, J. Long-range chromosomal interactions and gene regulation. Mol. Biosyst 4, 1046–1057 (2008).
Potts, P. R., Porteus, M. H. & Yu, H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 25, 3377–3388 (2006).
Strom, L., Lindroos, H. B., Shirahige, K. & Sjogren, C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell 16, 1003–1015 (2004).
Sjogren, C. & Nasmyth, K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr. Biol. 11, 991–995 (2001).
Heidinger-Pauli, J. M., Unal, E., Guacci, V. & Koshland, D. The kleisin subunit of cohesin dictates damage-induced cohesion. Mol. Cell 31, 47–56 (2008).
Unal, E. et al. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16, 991–1002 (2004).
Kim, B. J. et al. Genome-wide reinforcement of cohesin binding at pre-existing cohesin sites in response to ionizing radiation in human cells. J. Biol. Chem. 285, 22784–22792 (2010).
Yazdi, P. T. et al. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev. 16, 571–582 (2002).
Klein, F. et al. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98, 91–103 (1999).
Nasmyth, K., Peters, J. M. & Uhlmann, F. Splitting the chromosome: cutting the ties that bind sister chromatids. Novartis Found. Symp. 237, 113–133 (2001).
Doll, E. et al. Cohesin and recombination proteins influence the G1-to-S. transition in azygotic meiosis in Schizosaccharomyces pombe. Genetics 180, 727–740 (2008).
Bishop, A. J. & Schiestl, R. H. Role of homologous recombination in carcinogenesis. Exp. Mol. Pathol. 74, 94–105 (2003).
Reliene, R., Bishop, A. J. & Schiestl, R. H. Involvement of homologous recombination in carcinogenesis. Adv. Genet. 58, 67–87 (2007).
Moynahan, M. E. & Jasin, M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nature Rev. Mol. Cell Biol. 11, 196–207 (2010).
Modesti, M. & Kanaar, R. Homologous recombination: from model organisms to human disease. Genome Biol. 2, 1014.1–104.5 (2001).
Helleday, T., Petermann, E., Lundin, C., Hodgson, B. & Sharma, R. A. DNA repair pathways as targets for cancer therapy. Nature Rev. Cancer 8, 193–204 (2008).
Park, M. S. Expression of human RAD52 confers resistance to ionizing radiation in mammalian cells. J. Biol. Chem. 270, 15467–15470 (1995).
Husain, A., He, G., Venkatraman, E. S. & Spriggs, D. R. BRCA1 up-regulation is associated with repair-mediated resistance to cis-diamminedichloroplatinum(II). Cancer Res. 58, 1120–1123 (1998).
Bristow, R. G. & Hill, R. P. Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nature Rev. Cancer 8, 180–192 (2008).
Evers, B., Helleday, T. & Jonkers, J. Targeting homologous recombination repair defects in cancer. Trends Pharmacol. Sci. 31, 372–380 (2010).
Losada, A. & Hirano, T. Biology in pictures. New light on sticky sisters. Curr. Biol. 10, R615 (2000).
Nagao, K., Adachi, Y. & Yanagida, M. Separase-mediated cleavage of cohesin at interphase is required for DNA repair. Nature 430, 1044–1048 (2004).
Wang, Y. et al. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 14, 927–939 (2000).
Kim, J. S., Krasieva, T. B., LaMorte, V., Taylor, A. M. & Yokomori, K. Specific recruitment of human cohesin to laser-induced DNA damage. J. Biol. Chem. 277, 45149–45153 (2002).
van den Bosch, M., Bree, R. T. & Lowndes, N. F. The MRN complex: coordinating and mediating the response to broken chromosomes. EMBO Rep. 4, 844–849 (2003).
Ball, A. R., Jr. & Yokomori, K. Damage-induced reactivation of cohesin in postreplicative DNA repair. Bioessays 30, 5–9 (2008).
Stursberg, S., Riwar, B. & Jessberger, R. Cloning and characterization of mammalian SMC1 and SMC3 genes and proteins, components of the DNA recombination complexes RC-1. Gene 228, 1–12 (1999).
Warren, W. D., Lin, E., Nheu, T. V., Hime, G. R. & McKay, M. J. Drad21, a Drosophila rad21 homologue expressed in S-phase cells. Gene 250, 77–84 (2000).
Yan, J. et al. Mutational and genotype-phenotype correlation analyses in 28 Polish patients with Cornelia de Lange syndrome. Am. J. Med. Genet. A 140, 1531–1541 (2006).
Sehl, M. E. et al. Associations between single nucleotide polymorphisms in double-stranded DNA repair pathway genes and familial breast cancer. Clin. Cancer Res. 15, 2192–2203 (2009).
Ghiselli, G., Coffee, N., Munnery, C. E., Koratkar, R. & Siracusa, L. D. The cohesin SMC3 is a target the for beta-catenin/TCF4 transactivation pathway. J. Biol. Chem. 278, 20259–20267 (2003).
Acknowledgements
The authors would like to thank R. Ramsay and M.-C. Vozenin for comments on the manuscript. H.X. and M.J.M. were supported by grants from the Australian National Health and Medical Research Council (Project grants 400207, 400310 and 1007659) and the Cancer Council Victoria (091224).
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Rights and permissions
About this article
Cite this article
Xu, H., Tomaszewski, J. & McKay, M. Can corruption of chromosome cohesion create a conduit to cancer?. Nat Rev Cancer 11, 199–210 (2011). https://doi.org/10.1038/nrc3018
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc3018
This article is cited by
-
KNTC1 as a putative tumor oncogene in pancreatic cancer
Journal of Cancer Research and Clinical Oncology (2023)
-
miR-122 Inhibits the Cervical Cancer Development by Targeting the Oncogene RAD21
Biochemical Genetics (2022)
-
Conditional transcriptional relationships may serve as cancer prognostic markers
BMC Medical Genomics (2021)
-
Enhanced expression of cohesin loading factor NIPBL confers poor prognosis and chemotherapy resistance in non-small cell lung cancer
Journal of Translational Medicine (2015)
-
The AtRAD21.1 and AtRAD21.3 Arabidopsis cohesins play a synergistic role in somatic DNA double strand break damage repair
BMC Plant Biology (2014)