Abstract
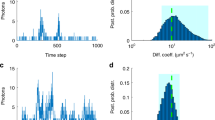
Probing transcription factor (TF)–DNA interactions remains challenging in complex in vivo systems such as mammalian embryos, especially when TF copy numbers and fluorescence background are high. To address this difficulty, fluorescence correlation spectroscopy (FCS) can be combined with the use of photoactivatable fluorescent proteins to achieve selective photoactivation of a subset of tagged TF molecules. This approach, termed paFCS, enables FCS measurements within single cell nuclei inside live embryos, and obtains autocorrelation data of a quality previously only attainable in simpler in vitro cell culture systems. Here, we present a protocol demonstrating the applicability of paFCS in developing mouse embryos by outlining its implementation on a commercial laser-scanning microscope. We also provide procedures for optimizing the photoactivation and acquisition parameters and determining key parameters describing TF–DNA binding. The entire procedure can be performed within ∼2 d (excluding embryo culture time), although the acquisition of each paFCS data set takes only ∼10 min. This protocol can be used to noninvasively reveal cell-to-cell variation in TF dynamics, as well as critical, fate-predicting changes over the course of early embryonic development.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
McNally, J.G., Müller, W.G., Walker, D., Wolford, R. & Hager, G.L. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287, 1262–1265 (2000).
Gebhardt, J.C.M. et al. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat. Methods 10, 421–426 (2013).
Mueller, F., Wach, P. & McNally, J.G. Evidence for a common mode of transcription factor interaction with chromatin as revealed by improved quantitative fluorescence recovery after photobleaching. Biophys. J. 94, 3323–3339 (2008).
Morisaki, T., Müller, W.G., Golob, N., Mazza, D. & McNally, J.G. Single-molecule analysis of transcription factor binding at transcription sites in live cells. Nat. Commun. 5, 4456 (2014).
Chen, J. et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 156, 1274–1285 (2014).
Mazza, D., Abernathy, A., Golob, N., Morisaki, T. & McNally, J.G. A benchmark for chromatin binding measurements in live cells. Nucleic Acids Res. 40, e119 (2012).
Sugo, N. et al. Single-molecule imaging reveals dynamics of CREB transcription factor bound to its target sequence. Sci. Rep. 5, 10662 (2015).
Speil, J. et al. Activated STAT1 transcription factors conduct distinct saltatory movements in the cell nucleus. Biophys. J. 101, 2592–2600 (2011).
Liu, Z. et al. 3D imaging of Sox2 enhancer clusters in embryonic stem cells. eLife 3, e04236 (2014).
Izeddin, I. et al. Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. eLife 3, e02230 (2014).
Zhao, Z.W., White, M.D., Bissiere, S., Levi, V. & Plachta, N. Quantitative imaging of mammalian transcriptional dynamics: from single cells to whole embryos. BMC Biol. 14, 115 (2016).
Hager, G.L., McNally, J.G. & Misteli, T. Transcription dynamics. Mol. Cell 35, 741–753 (2009).
Voss, T.C. & Hager, G.L. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat. Rev. Genet. 15, 69–81 (2014).
Coleman, R.A. et al. Imaging transcription: past, present, and future. Cold Spring Harb. Symp. Quant. Biol. 80, 1–8 (2015).
Darzacq, X. et al. Imaging transcription in living cells. Annu. Rev. Biophys. 38, 173–196 (2009).
Magde, D., Elson, E. & Webb, W.W. Thermodynamic fluctuations in a reacting system-measurement by fluorescence correlation spectroscopy. Phys. Rev. Lett. 29, 705–708 (1972).
Elson, E.L. Fluorescence correlation spectroscopy: past, present, future. Biophys. J. 101, 2855–2870 (2011).
Macháň, R. & Wohland, T. Recent applications of fluorescence correlation spectroscopy in live systems. FEBS Lett. 588, 3571–3584 (2014).
Digman, M.A. & Gratton, E. Lessons in fluctuation correlation spectroscopy. Annu. Rev. Phys. Chem. 62, 645–668 (2011).
Weidemann, T., Mücksch, J. & Schwille, P. Fluorescence fluctuation microscopy: a diversified arsenal of methods to investigate molecular dynamics inside cells. Curr. Opin. Struct. Biol. 28, 69–76 (2014).
Bacia, K. & Schwille, P. Practical guidelines for dual-color fluorescence cross-correlation spectroscopy. Nat. Protoc. 2, 2842–2856 (2007).
Krieger, J.W. et al. Imaging fluorescence (cross-) correlation spectroscopy in live cells and organisms. Nat. Protoc. 10, 1948–1974 (2015).
Rossow, M.J., Sasaki, J.M., Digman, M.A. & Gratton, E. Raster image correlation spectroscopy in live cells. Nat. Protoc. 5, 1761–1774 (2010).
Weidemann, T. Application of fluorescence correlation spectroscopy (FCS) to measure the dynamics of fluorescent proteins in living cells. in Fluorescence Spectroscopy and Microscopy: Methods and Protocols (eds. Engelborghs, Yves & Visser, Antonie J.W.G.) 539–555 (Humana Press, 2014).
Biggin, M.D. Animal transcription networks as highly connected, quantitative continua. Dev. Cell 21, 611–626 (2011).
Patterson, G.H. & Lippincott-Schwartz, J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science 297, 1873–1877 (2002).
Kaur, G. et al. Probing transcription factor diffusion dynamics in the living mammalian embryo with photoactivatable fluorescence correlation spectroscopy. Nat. Commun. 4, 1637 (2013).
White, M.D. et al. Long-lived binding of Sox2 to DNA predicts cell fate in the four-cell mouse embryo. Cell 165, 75–87 (2016).
Zhao, Z.W., Gebhardt, J.C.M., Suter, D.M. & Xie, X.S. Reply to 'Convergence of chromatin binding estimates in live cells'. Nat. Methods 10, 692 (2013).
Wang, Y., Wang, X., Wohland, T. & Sampath, K. Extracellular interactions and ligand degradation shape the nodal morphogen gradient. eLife 5, e13879 (2016).
Yu, S.R. et al. Fgf8 morphogen gradient forms by a source-sink mechanism with freely diffusing molecules. Nature 461, 533–536 (2009).
Shi, X. et al. Determination of dissociation constants in living zebrafish embryos with single wavelength fluorescence cross-correlation spectroscopy. Biophys. J. 97, 678–686 (2009).
Capoulade, J., Wachsmuth, M., Hufnagel, L. & Knop, M. Quantitative fluorescence imaging of protein diffusion and interaction in living cells. Nat. Biotech. 29, 835–839 (2011).
Wohland, T., Shi, X., Sankaran, J. & Stelzer, E.H.K. Single plane illumination fluorescence correlation spectroscopy (SPIM-FCS) probes inhomogeneous three-dimensional environments. Opt. Express 18, 10627–10641 (2010).
Digman, M.A. et al. Measuring fast dynamics in solutions and cells with a laser scanning microscope. Biophys. J. 89, 1317–1327 (2005).
Digman, M.A. & Gratton, E. Imaging barriers to diffusion by pair correlation functions. Biophys. J. 97, 665–673 (2009).
Ries, J., Chiantia, S. & Schwille, P. Accurate determination of membrane dynamics with line-scan FCS. Biophys. J. 96, 1999–2008 (2009).
Pantazis, P. & González-Gaitán, M. Localized multiphoton photoactivation of paGFP in Drosophila wing imaginal discs. J. Biomed. Opt. 12, 044004 (2007).
Plachta, N., Bollenbach, T., Pease, S., Fraser, S.E. & Pantazis, P. Oct4 kinetics predict cell lineage patterning in the early mammalian embryo. Nat. Cell Biol. 13, 117–123 (2011).
Wang, S., Moffitt, J.R., Dempsey, G.T., Xie, X.S. & Zhuang, X. Characterization and development of photoactivatable fluorescent proteins for single-molecule–based superresolution imaging. Proc. Natl. Acad. Sci. USA 111, 8452–8457 (2014).
Mohr, M.A., Argast, P. & Pantazis, P. Labeling cellular structures in vivo using confined primed conversion of photoconvertible fluorescent proteins. Nat. Protoc. 11, 2419–2431 (2016).
Rigler, R., Mets, Ü., Widengren, J. & Kask, P. Fluorescence correlation spectroscopy with high count rate and low background: analysis of translational diffusion. Eur. Biophys. J. 22, 169–175 (1993).
Widengren, J., Mets, Ü. & Rigler, R. Photodynamic properties of green fluorescent proteins investigated by fluorescence correlation spectroscopy. Chem. Phys. 250, 171–186 (1999).
Halford, S.E. & Marko, J.F. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 32, 3040–3052 (2004).
Michelman-Ribeiro, A. et al. Direct measurement of association and dissociation rates of DNA binding in live cells by fluorescence correlation spectroscopy. Biophys. J. 97, 337–346 (2009).
Elson, E.L. Brief introduction to fluorescence correlation spectroscopy. in Methods in Enzymology Vol. 518 (ed. Tetin, S.Y.), 11–41 (Academic Press, 2013).
Sobell, H.M. Actinomycin and DNA transcription. Proc. Natl. Acad. Sci. USA 82, 5328–5331 (1985).
Yoshida, M., Horinouchi, S. & Beppu, T. Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. BioEssays 17, 423–430 (1995).
Behringer, R., Gertsenstein, M., Nagy, K.V. & Nagy, A. Manipulating the Mouse Embryo: A Laboratory Manual 4th ed. (Cold Spring Harbor Laboratory Press, 2014).
Müller, P., Schwille, P. & Weidemann, T. PyCorrFit—generic data evaluation for fluorescence correlation spectroscopy. Bioinformatics 30, 2532–2533 (2014).
Ries, J. et al. Automated suppression of sample-related artifacts in fluorescence correlation spectroscopy. Opt. Express 18, 11073–11082 (2010).
Beechem, J.M. Global analysis of biochemical and biophysical data. in Methods Enzymol. Vol. 210, 37–54 (Academic Press, 1992).
Wachsmuth, M., Waldeck, W. & Langowski, J. Anomalous diffusion of fluorescent probes inside living cell nuclei investigated by spatially-resolved fluorescence correlation spectroscopy. J. Mol. Biol. 298, 677–689 (2000).
He, J., Guo, S.-M. & Bathe, M. Bayesian approach to the analysis of fluorescence correlation spectroscopy data I: theory. Anal. Chem. 84, 3871–3879 (2012).
Sun, G. et al. Bayesian model selection applied to the analysis of fluorescence correlation spectroscopy data of fluorescent proteins in vitro and in vivo. Anal. Chem. 87, 4326–4333 (2015).
Schwille, P., Bieschke, J. & Oehlenschläger, F. Kinetic investigations by fluorescence correlation spectroscopy: the analytical and diagnostic potential of diffusion studies. Biophys. Chem. 66, 211–228 (1997).
Acknowledgements
We thank J. Silva for help with embryo isolation and microinjection, and V. Levi for advice on modeling. This work was supported by an A*STAR National Science Scholarship (to Z.W.Z.), a Human Frontiers Science Program Fellowship (to J.Z.), and an A*STAR Investigatorship and EMBO Young Investigator grants (to N.P.).
Author information
Authors and Affiliations
Contributions
N.P. conceived the project; Z.W.Z. and Y.D.A. performed the microscope calibration, optimization of photoactivation/acquisition parameters, and paFCS measurements; M.D.W. and J.Z. performed drug treatment and mutant TF experiments. Y.D.A. performed data analysis and modeling. S.B. performed embryo isolation, culture, and mRNA microinjections. Z.W.Z., M.D.W., Y.D.A., J.Z., S.B. and N.P. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Zhao, Z., White, M., Alvarez, Y. et al. Quantifying transcription factor–DNA binding in single cells in vivo with photoactivatable fluorescence correlation spectroscopy. Nat Protoc 12, 1458–1471 (2017). https://doi.org/10.1038/nprot.2017.051
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2017.051
This article is cited by
-
From whole-mount to single-cell spatial assessment of gene expression in 3D
Communications Biology (2020)
-
Using enhanced number and brightness to measure protein oligomerization dynamics in live cells
Nature Protocols (2019)
-
Understanding biochemical processes in the presence of sub-diffusive behavior of biomolecules in solution and living cells
Biophysical Reviews (2019)
-
Optimal fluorescent protein tags for quantifying protein oligomerization in living cells
Scientific Reports (2018)
-
Monomerization of the photoconvertible fluorescent protein SAASoti by rational mutagenesis of single amino acids
Scientific Reports (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.