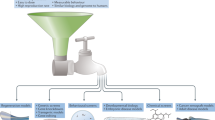
Abstract
The zebrafish has emerged as an important model for whole-organism small-molecule screening. However, most zebrafish-based chemical screens have achieved only mid-throughput rates. Here we describe a versatile whole-organism drug discovery platform that can achieve true high-throughput screening (HTS) capacities. This system combines our automated reporter quantification in vivo (ARQiv) system with customized robotics, and is termed 'ARQiv-HTS'. We detail the process of establishing and implementing ARQiv-HTS: (i) assay design and optimization, (ii) calculation of sample size and hit criteria, (iii) large-scale egg production, (iv) automated compound titration, (v) dispensing of embryos into microtiter plates, and (vi) reporter quantification. We also outline what we see as best practice strategies for leveraging the power of ARQiv-HTS for zebrafish-based drug discovery, and address technical challenges of applying zebrafish to large-scale chemical screens. Finally, we provide a detailed protocol for a recently completed inaugural ARQiv-HTS effort, which involved the identification of compounds that elevate insulin reporter activity. Compounds that increased the number of insulin-producing pancreatic beta cells represent potential new therapeutics for diabetic patients. For this effort, individual screening sessions took 1 week to conclude, and sessions were performed iteratively approximately every other day to increase throughput. At the conclusion of the screen, more than a half million drug-treated larvae had been evaluated. Beyond this initial example, however, the ARQiv-HTS platform is adaptable to almost any reporter-based assay designed to evaluate the effects of chemical compounds in living small-animal models. ARQiv-HTS thus enables large-scale whole-organism drug discovery for a variety of model species and from numerous disease-oriented perspectives.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Swinney, D.C. & Anthony, J. How were new medicines discovered? Nat. Rev. Drug Discov. 10, 507–519 (2011).
Eder, A. et al. Effects of proarrhythmic drugs on relaxation time and beating pattern in rat engineered heart tissue. Basic Res. Cardiol. 109, 436 (2014).
Walker, S.L. et al. Automated reporter quantification in vivo: high-throughput screening method for reporter-based assays in zebrafish. PLoS One 7, e29916 (2012).
Inglese, J. et al. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc. Natl. Acad. Sci. USA 103, 11473–11478 (2006).
Wang, G. et al. First quantitative high-throughput screen in zebrafish identifies novel pathways for increasing pancreatic β-cell mass. Elife 4 http://dx.doi.org/10.7554/eLife.08261 (2015).
Scannell, J.W., Blanckley, A., Boldon, H. & Warrington, B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat. Rev. Drug Discov. 11, 191–200 (2012).
Rishton, G.M. Nonleadlikeness and leadlikeness in biochemical screening. Drug Discov. Today 8, 86–96 (2003).
Medina, O., Estrada, J.C. & Servin, M. Robust adaptive phase-shifting demodulation for testing moving wavefronts. Opt. Express 21, 29687–29694 (2013).
Walker, M.J.A., Barrett, T. & Guppy, L.J. Functional pharmacology: the drug discovery bottleneck? Drug Discov. Today TARGETS 3, 208–215 (2004).
Rennekamp, A.J. & Peterson, R.T. From phenotype to mechanism after zebrafish small molecule screens. Drug Discov. Today. Dis. Models 10, e51–e55 (2013).
Rennekamp, A.J. & Peterson, R.T. 15 years of zebrafish chemical screening. Curr. Opin. Chem. Biol. 24, 58–70 (2015).
Peterson, R.T., Link, B.A., Dowling, J.E. & Schreiber, S.L. Small molecule developmental screens reveal the logic and timing of vertebrate development. Proc. Natl. Acad. Sci. USA 97, 12965–12969 (2000).
Peterson, R.T. et al. Chemical suppression of a genetic mutation in a zebrafish model of aortic coarctation. Nat. Biotechnol. 22, 595–599 (2004).
Burns, C.G. et al. High-throughput assay for small molecules that modulate zebrafish embryonic heart rate. Nat. Chem. Biol. 1, 263–264 (2005).
Stern, H.M. et al. Small molecules that delay S phase suppress a zebrafish bmyb mutant. Nat. Chem. Biol. 1, 366–370 (2005).
Murphey, R.D. & Zon, L.I. Small molecule screening in the zebrafish. Methods 39, 255–261 (2006).
Anderson, C. et al. Chemical genetics suggests a critical role for lysyl oxidase in zebrafish notochord morphogenesis. Mol. Biosyst. 3, 51–59 (2007).
Molina, G.A., Watkins, S.C. & Tsang, M. Generation of FGF reporter transgenic zebrafish and their utility in chemical screens. BMC Dev. Biol. 7, 62 (2007).
Owens, K.N. et al. Identification of genetic and chemical modulators of zebrafish mechanosensory hair cell death. PLoS Genet. 4, e1000020 (2008).
Cao, Y. et al. Chemical modifier screen identifies HDAC inhibitors as suppressors of PKD models. Proc. Natl. Acad. Sci. USA 106, 21819–21824 (2009).
Molina, G. et al. Zebrafish chemical screening reveals an inhibitor of Dusp6 that expands cardiac cell lineages. Nat. Chem. Biol. 5, 680–687 (2009).
Yeh, H.H. et al. Ha-ras oncogene-induced Stat3 phosphorylation enhances oncogenicity of the cell. DNA Cell Biol. 28, 131–139 (2009).
Coffin, A.B. et al. Chemical screening for hair cell loss and protection in the zebrafish lateral line. Zebrafish 7, 3–11 (2010).
de Groh, E.D. et al. Inhibition of histone deacetylase expands the renal progenitor cell population. J. Am. Soc. Nephrol. 21, 794–802 (2010).
Kokel, D. et al. Rapid behavior-based identification of neuroactive small molecules in the zebrafish. Nat. Chem. Biol. 6, 231–237 (2010).
Paik, E.J., de Jong, J.L.O., Pugach, E., Opara, P. & Zon, L.I. A chemical genetic screen in zebrafish for pathways interacting with cdx4 in primitive hematopoiesis. Zebrafish 7, 61–68 (2010).
Rihel, J. et al. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science 327, 348–351 (2010).
Milan, D.J., Peterson, T.A., Ruskin, J.N., Peterson, R.T. & MacRae, C.A. Drugs that induce repolarization abnormalities cause bradycardia in zebrafish. Circulation 107, 1355–1358 (2003).
Ali, S., van Mil, H.G.J. & Richardson, M.K. Large-scale assessment of the zebrafish embryo as a possible predictive model in toxicity testing. PLoS One 6, e21076 (2011).
Langheinrich, U. Zebrafish: a new model on the pharmaceutical catwalk. Bioessays 25, 904–912 (2003).
North, T.E. et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature 447, 1007–1011 (2007).
Peravali, R. et al. Automated feature detection and imaging for high-resolution screening of zebrafish embryos. Biotechniques 50, 319–324 (2011).
MacRae, C.A. & Peterson, R.T. Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov. 14, 721–731 (2015).
Mathias, J.R., Saxena, M.T. & Mumm, J.S. Advances in zebrafish chemical screening technologies. Future Med. Chem. 4, 1811–1822 (2012).
Leung, C.K. et al. An ultra high-throughput, whole-animal screen for small molecule modulators of a specific genetic pathway in Caenorhabditis elegans. PLoS One 8, e62166 (2013).
Goktug, A.N., Chai, S.C. & Chen, T. Data analysis approaches in high throughput screening. Drug Discovery (ed. El-Shemy, H.A.). (InTech, 2013).
Zhang, X.D. et al. Integrating experimental and analytic approaches to improve data quality in genome-wide RNAi screens. J. Biomol. Screen. 13, 378–389 (2008).
Zhang, J., Chung, T. & Oldenburg, K. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4, 67–73 (1999).
Zhang, X.D. Novel analytic criteria and effective plate designs for quality control in genome-scale RNAi screens. J. Biomol. Screen. 13, 363–377 (2008).
Zhang, X.D. Illustration of SSMD, z score, SSMD*, z* score, and t statistic for hit selection in RNAi high-throughput screens. J. Biomol. Screen. 16, 775–785 (2011).
Graf, S.F., Hötzel, S., Liebel, U., Stemmer, A. & Knapp, H.F. Image-based fluidic sorting system for automated zebrafish egg sorting into multiwell plates. J. Lab. Autom. 16, 105–111 (2011).
Parsons, M.J. et al. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech. Dev. 126, 898–912 (2009).
Lawrence, M. & Lang, D. RGtk2: a graphical user interface toolkit for R. J. Stat. Softw. 37, 1–52 (2010).
Borchers, H.W. pracma: Practical Numerical Math Functions. (2015). https://rdrr.io/rforge/pracma/ (2016).
Rovira, M. et al. Chemical screen identifies FDA-approved drugs and target pathways that induce precocious pancreatic endocrine differentiation. Proc. Natl. Acad. Sci. USA 108, 19264–19269 (2011).
Ariga, J., Walker, S.L. & Mumm, J.S. Multicolor time-lapse imaging of transgenic zebrafish: visualizing retinal stem cells activated by targeted neuronal cell ablation. J. Vis. Exp. 43, e2093 (2010).
Halsey, L.G., Curran-Everett, D., Vowler, S.L. & Drummond, G.B. The fickle P value generates irreproducible results. Nat. Methods 12, 179–185 (2015).
Westerfield, M. A guide for the laboratory use of zebrafish (Danio rerio) (Eugene, OR: University of Oregon Press, 2007).
Adatto, I., Lawrence, C., Thompson, M. & Zon, L.I. A new system for the rapid collection of large numbers of developmentally staged zebrafish embryos. PLoS One 6, e21715 (2011).
Hallare, A., Nagel, K., Köhler, H.-R. & Triebskorn, R. Comparative embryotoxicity and proteotoxicity of three carrier solvents to zebrafish (Danio rerio) embryos. Ecotoxicol. Environ. Saf. 63, 378–388 (2006).
David, R.M. et al. Interference with xenobiotic metabolic activity by the commonly used vehicle solvents dimethylsulfoxide and methanol in zebrafish (Danio rerio) larvae but not Daphnia magna. Chemosphere 88, 912–917 (2012).
Zhang, X.D. A pair of new statistical parameters for quality control in RNA interference high-throughput screening assays. Genomics 89, 552–561 (2007).
Acknowledgements
We are grateful to R. L. Daily, 3rd (integration designer, Hudson Robotics) for creating the ARQiv-HTS workstation schematics. A huge thanks goes to our in vivo drug discovery collaborators J. Liu (Department of Pharmacology and Molecular Science, Johns Hopkins School of Medicine) and M. Parsons (Department of Surgery, Johns Hopkins School of Medicine) and their research teams. We also thank M. Saxena for her editorial assistance, as well as other past and present Mumm and Luminomics laboratory members for helpful discussions. This work was supported by grants to J.S.M. from the NIH (RC4DK090816, R01EY022810, R41TR000945, and F31EY021713), DoD (MR130301), and the Foundation Fighting Blindness (TA-NMT-0614-0643-JHU-WG).
Author information
Authors and Affiliations
Contributions
J.S.M. conceived of the ARQiv-HTS platform, designed reporter-based assay systems, and supervised the project. D.T.W., A.U.E., L.Z., S.S., S.K.R., and S.L.W. accomplished large-scale implementation of the ARQiv-HTS system and analyzed data. G.W., D.D., S.L.W., H.J., and J.Q. developed the software packages for assay optimization and real-time data analysis. D.T.W., A.U.E., and J.S.M. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
J.S.M. is a founder and stakeholder of Luminomics, a company that is collaborating with Hudson Robotics and Union Biometrica to develop a series of benchtop AQRiv-HTS assay platforms as commercial products.
Integrated supplementary information
Supplementary Figure 1 Data transformation.
A: Representative data from an ARQiv-HTS assay quality test with arbitrary fluorescent units (AFU) expressed on an arithmetic scale. The data show unequal variance between the negative (green) and positive (red) controls. B: Base 2 log transformation of the data shown in A. Log transformation achieves more symmetrically distributed data around the respective means, as in a normal distribution, and can also facilitate a larger dynamic range of the effect size metrics used to evaluate tested compounds (e.g., increased SSMD scores, compare upper right in A and B).
Supplementary Figure 2 Economic large-scale egg production units.
A: Top view of simple grouped breeding system assembled from plastic storage units showing the uppermost mating chamber (empty). Dashed boxes represent areas where inserted mesh screen allows eggs to pass through to a middle collection chamber; dashed circle shows area where micron mesh is inserted in the collection chamber to allow water drainage. B: Close-up side view of all three chambers, mating, collection, and water, seated one inside the other. Arrows indicate egg movement from mating to collection chamber. C: Grouped breeding unit in use, egg production is correlated to density of breeders in mating chamber, plastic ‘plants’ can be dropped in to further stimulate breeding. C’: Measurement of eggs with graduated cylinder (once settled, 500-600eggs/mL). D: Top view of empty drop-in mating chambers. E: Side view of drop-in mating chamber (in use). Arrows indicate egg movement from tank to collection chamber.
Supplementary Figure 3 ARQiv package graphical user interface (GUI).
Upon installing the ARQiv R-based package, the GUI above is available for simplifying ARQiv data processing. The use of this GUI is detailed within relevant sections of the protocol. Briefly, the ARQiv R package includes functions that fall into two categories - those applied to ‘Pre-screening Assay Optimization' (upper panel) and 'Compound Analysis' (lower panel). The functions allow the user to calculate background signal, determine sample size, run quality control tests, perform virtual experiments to simulate compound efficacy - and finally, to perform compound analysis during iterative drug screen cycles.
Supplementary Figure 4 Titration-based ARQiv-HTS assay diagram.
Compounds are tested at a total six concentrations at a sample number of 16 per compound concentration, thus one 96 well plate per compound (center 96 well plate). To account for the possibility of signal changes over time, positive and negative control plates (red and green, respectively) ‘bookend’ every set of 10 tested compound plates. This process is reiterated for each series of tested compounds.
Supplementary Figure 5 COPAS-based larval fish sorting and microtiter plate dispensing.
Sorting and dispensing of transgenic larvae into microtiter plates can be automated using the COPAS-XL system. Fish are illuminated with a 561 solid-state nm laser as they pass through a flow cell (analysis chamber) wherein they are gated/sorted based on extinction (i.e., size and internal structure of object), time of flight (i.e., length of object), and fluorescence (emission at 610 nm +/-10). Upper panels are ‘gating dot plots’ denoting extinction (Ext, y-axis) and time of flight (Tof, x-axis) parameters used for size-based sorting of fish/non-fish objects as determined by user-defined ‘gate region’ (e.g., interior to dashed line in upper panel). Lower panel is fluorescence-based sorting of transgenic fish via user-defined ‘gate region’ in ‘sorting dot plot’ denoting ‘RFP’ signal (Red, y-axis) and time of flight (Tof, x-axis). Red bracket represents transgenic fish sorted into wells.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 (PDF 573 kb)
Supplementary Data
zip file (ZIP 22 kb)
Rights and permissions
About this article
Cite this article
White, D., Eroglu, A., Wang, G. et al. ARQiv-HTS, a versatile whole-organism screening platform enabling in vivo drug discovery at high-throughput rates. Nat Protoc 11, 2432–2453 (2016). https://doi.org/10.1038/nprot.2016.142
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.142
This article is cited by
-
Long lasting anxiety following early life stress is dependent on glucocorticoid signaling in zebrafish
Scientific Reports (2022)
-
NTR 2.0: a rationally engineered prodrug-converting enzyme with substantially enhanced efficacy for targeted cell ablation
Nature Methods (2022)
-
The LipoGlo reporter system for sensitive and specific monitoring of atherogenic lipoproteins
Nature Communications (2019)
-
Functional missense and splicing variants in the retinoic acid catabolizing enzyme CYP26C1 in idiopathic short stature
European Journal of Human Genetics (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.