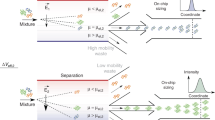
Abstract
Herein we describe a protocol that uses hollow-fiber flow field-flow fractionation (FFF) coupled with multiangle light scattering (MALS) for hydrodynamic size-based separation and characterization of complex protein aggregates. The fractionation method, which requires 1.5 h to run, was successfully modified from the analysis of protein aggregates, as found in simple protein mixtures, to complex aggregates, as found in total cell lysates. In contrast to other related methods (filter assay, analytical ultracentrifugation, gel electrophoresis and size-exclusion chromatography), hollow-fiber flow FFF coupled with MALS allows a flow-based fractionation of highly purified protein aggregates and simultaneous measurement of their molecular weight, r.m.s. radius and molecular conformation (e.g., round, rod-shaped, compact or relaxed). The polyethersulfone hollow fibers used, which have a 0.8-mm inner diameter, allow separation of as little as 20 μg of total cell lysates. In addition, the ability to run the samples in different denaturing and nondenaturing buffer allows defining true aggregates from artifacts, which can form during sample preparation. The protocol was set up using Paraquat-induced carbonylation, a model that induces protein aggregation in cultured cells. This technique will advance the biochemical, proteomic and biophysical characterization of molecular-weight aggregates associated with protein mutations, as found in many CNS degenerative diseases, or chronic oxidative stress, as found in aging, and chronic metabolic and inflammatory conditions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kopito, R.R. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10, 524–530 (2000).
Kalia, L.V., Kalia, S.K., McLean, P.J., Lozano, A.M. & Lang, A.E. α-Synuclein oligomers and clinical implications for Parkinson disease. Ann. Neurol. 73, 155–169 (2013).
Wang, Y. et al. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum. Mol. Genet. 18, 4153–4170 (2009).
Perlmutter, D.H. The role of autophagy in α-1-antitrypsin deficiency: a specific cellular response in genetic diseases associated with aggregation-prone proteins. Autophagy 2, 258–263 (2006).
Fink, A.L. The aggregation and fibrillation of α-synuclein. Acc. Chem. Res. 39, 628–634 (2006).
Cannizzo, E.S. et al. Age-related oxidative stress compromises endosomal proteostasis. Cell Rep. 2, 136–149 (2012).
Cannizzo, E.S., Clement, C.C., Sahu, R., Follo, C. & Santambrogio, L. Oxidative stress, inflamm-aging and immunosenescence. J. Proteomics 74, 2313–2323 (2011).
Scharf, B. et al. Age-related carbonylation of fibrocartilage structural proteins drives tissue degenerative modification. Chem. Biol. 20, 922–934 (2013).
Bence, N.F., Sampat, R.M. & Kopito, R.R. Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292, 1552–1555 (2001).
Berke, S.J. & Paulson, H.L. Protein aggregation and the ubiquitin proteasome pathway: gaining the UPPer hand on neurodegeneration. Curr. Opin. Genet. Dev. 13, 253–261 (2003).
Cuervo, A.M., Wong, E.S. & Martinez-Vicente, M. Protein degradation, aggregation, and misfolding. Mov. Disord. 25 (suppl. 1), S49–S54 (2010).
Holmberg, M. & Nollen, E.A. Analyzing modifiers of protein aggregation in C. elegans by native agarose gel electrophoresis. Methods Mol. Biol. 1017, 193–199 (2013).
Zhang, Y. & Calderwood, S.K. Autophagy, protein aggregation and hyperthermia: a mini-review. Int. J. Hyperthermia 27, 409–414 (2011).
Liu, C., Gao, Y., Barrett, J. & Hu, B. Autophagy and protein aggregation after brain ischemia. J. Neurochem. 115, 68–78 (2010).
Lok, C.N., Sy, L.K., Liu, F. & Che, C.M. Activation of autophagy of aggregation-prone ubiquitinated proteins by timosaponin A-III. J. Biol. Chem. 286, 31684–31696 (2011).
Reschiglian, P. & Moon, M.H. Flow field-flow fractionation: a pre-analytical method for proteomics. J. Proteomics 71, 265–276 (2008).
Giddings, J.C. A new separation concept based on a coupling of concentration and flow nonuniformities. Separ. Sci. 1, 123–125 (1966).
Giddings, J.C. The conceptual basis of field-flow fractionation. J. Chem. Educ. 50, 667–669 (1973).
Lee, J.Y., Min, H.K., Choi, D. & Moon, M.H. Profiling of phospholipids in lipoproteins by multiplexed hollow fiber flow field-flow fractionation and nanoflow liquid chromatography–tandem mass spectrometry. J. Chromatogr. A 1217, 1660–1666 (2010).
Reschiglian, P., Zattoni, A., Roda, B. & Cinque, L. On-line hollow-fiber flow field-flow fractionation-electrospray ionization/time-of-flight mass spectrometry of intact proteins. Anal. Chem. 77, 47–56 (2005).
Oueslati, A., Fournier, M. & Lashuel, H.A. Role of post-translational modifications in modulating the structure, function and toxicity of α-synuclein: implications for Parkinson's disease pathogenesis and therapies. Prog. Brain Res. 183, 115–145 (2010).
Nixon, R.A. The role of autophagy in neurodegenerative disease. Nat. Med. 19, 983–997 (2013).
Cuervo, A.M., Stefanis, L., Fredenburg, R., Lansbury, P.T. & Sulzer, D. Impaired degradation of mutant α-synuclein by chaperone-mediated autophagy. Science 305, 1292–1295 (2004).
Lee, S.J. et al. A detergent-insoluble membrane compartment contains Aβ in vivo. Nat. Med. 4, 730–734 (1998).
Lemere, C.A. et al. The E280A presenilin 1 Alzheimer mutation produces increased Aβ 42 deposition and severe cerebellar pathology. Nat. Med. 2, 1146–1150 (1996).
Phiel, C.J., Wilson, C.A., Lee, V.M. & Klein, P.S. GSK-3α regulates production of Alzheimer's disease amyloid-β peptides. Nature 423, 435–439 (2003).
Wong, E.S. et al. Autophagy-mediated clearance of aggresomes is not a universal phenomenon. Hum. Mol. Genet. 17, 2570–2582 (2008).
Dehay, B. & Bertolotti, A. Critical role of the proline-rich region in huntingtin for aggregation and cytotoxicity in yeast. J. Biol. Chem. 281, 35608–35615 (2006).
DiFiglia, M. et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277, 1990–1993 (1997).
Fiumara, F., Fioriti, L., Kandel, E.R. & Hendrickson, W.A. Essential role of coiled coils for aggregation and activity of Q/N-rich prions and polyQ proteins. Cell 143, 1121–1135 (2010).
Arena, S., Salzano, A.M., Renzone, G., D'Ambrosio, C. & Scaloni, A. Non-enzymatic glycation and glycoxidation protein products in foods and diseases: an interconnected, complex scenario fully open to innovative proteomic studies. Mass Spectrom. Rev. 33, 49–77 (2014).
Gillery, P. Oxidative stress and protein glycation in diabetes mellitus. Ann. Biol. Clin. (Paris) 64, 309–314 (2006).
Lapolla, A., Molin, L. & Traldi, P. Protein glycation in diabetes as determined by mass spectrometry. Int. J. Endocrinol. 2013, 412103 (2013).
Perkins, B.A. et al. Serum levels of advanced glycation endproducts and other markers of protein damage in early diabetic nephropathy in type 1 diabetes. PLoS ONE 7, e35655 (2012).
Simm, A. Protein glycation during aging and in cardiovascular disease. J. Proteomics 92, 248–259 (2013).
Janue, A., Olive, M. & Ferrer, I. Oxidative stress in desminopathies and myotilinopathies: a link between oxidative damage and abnormal protein aggregation. Brain Pathol. 17, 377–388 (2007).
Takalo, M., Salminen, A., Soininen, H., Hiltunen, M. & Haapasalo, A. Protein aggregation and degradation mechanisms in neurodegenerative diseases. Am. J. Neurodegener. Dis. 2, 1–14 (2013).
Shelkovnikova, T.A. et al. Proteinopathies—forms of neurodegenerative disorders with protein aggregation-based pathology. Mol. Biol. (Mosk) 46, 402–415 (2012).
Riley, B.E. et al. Ubiquitin accumulation in autophagy-deficient mice is dependent on the Nrf2-mediated stress response pathway: a potential role for protein aggregation in autophagic substrate selection. J. Cell Biol. 191, 537–552 (2010).
Tyedmers, J., Mogk, A. & Bukau, B. Cellular strategies for controlling protein aggregation. Nat. Rev. Mol. Cell Biol. 11, 777–788 (2010).
Cummings, C.J. et al. Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat. Genet. 19, 148–154 (1998).
Demasi, M. & Davies, K.J. Proteasome inhibitors induce intracellular protein aggregation and cell death by an oxygen-dependent mechanism. FEBS Lett. 542, 89–94 (2003).
Lu, M., Echeverri, F. & Moyer, B.D. Endoplasmic reticulum retention, degradation, and aggregation of olfactory G-protein coupled receptors. Traffic 4, 416–433 (2003).
Wyttenbach, A. et al. Effects of heat shock, heat shock protein 40 (HDJ-2), and proteasome inhibition on protein aggregation in cellular models of Huntington's disease. Proc. Natl. Acad. Sci. USA 97, 2898–2903 (2000).
Reschiglian, P. et al. Hollow-fiber flow field-flow fractionation with multi-angle laser scattering detection for aggregation studies of therapeutic proteins. Anal. Bioanal. Chem. 406, 1619–1627 (2014).
Kang, D. & Moon, M.H. Hollow fiber flow field-flow fractionation of proteins using a microbore channel. Anal. Chem. 77, 4207–4212 (2005).
Johann, C. et al. A novel approach to improve operation and performance in flow field-flow fractionation. J. Chromatogr. A 1218, 4126–4131 (2011).
Park, I., Paeng, K.-J., Kang, D. & Moon, M.H. Performance of hollow-fiber flow field-flow fractionation in protein separation. J. Separ. Sci. 28, 2043–2049 (2005).
Zattoni, A. et al. Hollow-fiber flow field-flow fractionation of whole blood serum. J. Chromatogr. A 1183, 135–142 (2008).
Reschiglian, P., Zattoni, A., Rambaldi, D.C., Roda, A. & Moon, M.H. Hollow-fiber flow field-flow fractionation for mass spectrometry: from proteins to whole bacteria. In Detection of Biological Agents for the Prevention of Bioterrorism (ed. Banoub, J.) 13–36 (Springer, 2011).
Zattoni, A., Rambaldi, D.C., Casolari, S., Roda, B. & Reschiglian, P. Tandem hollow-fiber flow field-flow fractionation. J. Chromatogr. A 1218, 4132–4137 (2011).
Lee, J.Y., Kim, K.H. & Moon, M.H. Evaluation of multiplexed hollow fiber flow field-flow fractionation for semi-preparative purposes. J. Chromatogr. A 1216, 6539–6542 (2009).
Kang, D., Ji, E.S., Moon, M.H. & Yoo, J.S. Lectin-based enrichment method for glycoproteomics using hollow fiber flow field-flow fractionation: application to Streptococcus pyogenes. J. Proteome Res. 9, 2855–2862 (2010).
Reschiglian, P., Zattoni, A., Cinque, L. & Roda, B. Hollow-fiber flow field-flow fractionation for whole bacteria analysis by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 76, 2103–2111 (2004).
Kang, D. & Moon, M.H. Development of non-gel-based two-dimensional separation of intact proteins by an on-line hyphenation of capillary isoelectric focusing and hollow fiber flow field-flow fractionation. Anal. Chem. 78, 5789–5798 (2006).
Silveira, J.R. et al. The most infectious prion protein particles. Nature 437, 257–261 (2005).
Stegemann, J., Ventzki, R., Schrödel, A. & de Marco, A. Comparative analysis of protein aggregates by blue native electrophoresis and subsequent sodium dodecyl sulfate-polyacrylamide gel electrophoresis in a three-dimensional geometry gel. Proteomics 5, 2002–2009 (2005).
Ishii, T. et al. Characterization of acrolein-induced protein cross-links. Free Radic. Res. 41, 1253–1260 (2007).
Linetsky, M., Shipova, E., Cheng, R. & Ortwerth, B.J. Glycation by ascorbic acid oxidation products leads to the aggregation of lens proteins. Biochim. Biophys. Acta 1782, 22–34 (2008).
Scharf, B. et al. Age-related carbonylation of fibrocartilage structural proteins drives tissue degenerative modification. Chem. Biol. 20, 922–934 (2013).
Mirzaei, H. & Regnier, F. Protein:protein aggregation induced by protein oxidation. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 873, 8–14 (2008).
Carpenter, J.F. et al. Potential inaccurate quantitation and sizing of protein aggregates by size-exclusion chromatography: essential need to use orthogonal methods to assure the quality of therapeutic protein products. J. Pharm. Sci. 99, 2200–2208 (2010).
Arakawa, T., Ejima, D., Li, T. & Philo, J.S. The critical role of mobile phase composition in size exclusion chromatography of protein pharmaceuticals. J. Pharm. Sci. 99, 1674–1692 (2010).
Kawahara, K. & Tanford, C. Viscosity and density of aqueous solutions of urea and guanidine hydrochloride. J. Biol. Chem. 241, 3228–3232 (1966).
Acknowledgements
We thank Superon for the loan of the Eclipse DUALTEC flow FFF separation system and Eclipse ISIS software, and Wyatt Technology for the loan of the MALS detector DAWN EOS and Astra software. We thank C. Johann for the valuable suggestions and comments on the manuscript and S. Elsenberg (Superon) for technical assistance. The work was supported by National Institute on Aging (NIA) grant PO1AG031781 to A.M.C. and PO1AG031782 to L.S. V.Z. is supported by the PhD program in Genetics and Cell Biology at the University of Tuscia, Department of Ecology and Biology (DEB).
Author information
Authors and Affiliations
Contributions
M.T., B.R., A.Z., P.R., A.M.C. and L.S. designed the experiments; M.T., V.Z., C.C.C., F.B., A.M.U. and J.A.R.-N. performed the experiments; and M.T., V.Z., C.C.C., B.R., A.Z., P.R., A.M.C. and L.S. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
A.Z., B.R. and P.R. are associates of the academic spinoff company byFlow SRL. The company mission includes know-how transfer, development, and application of novel technologies and methodologies for the analysis and characterization of samples of nano-biotechnological interest.
Rights and permissions
About this article
Cite this article
Tanase, M., Zolla, V., Clement, C. et al. Hydrodynamic size-based separation and characterization of protein aggregates from total cell lysates. Nat Protoc 10, 134–148 (2015). https://doi.org/10.1038/nprot.2015.009
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2015.009
This article is cited by
-
Characterising protein/detergent complexes by triple-detection size-exclusion chromatography
Biological Procedures Online (2016)
-
Role of Carbonyl Modifications on Aging-Associated Protein Aggregation
Scientific Reports (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.