Abstract
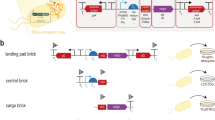
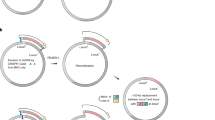
Conjugative assembly genome engineering (CAGE) is a precise method of genome assembly using conjugation to hierarchically combine distinct genotypes from multiple Escherichia coli strains into a single chimeric genome. CAGE permits large-scale transfer of specified genomic regions between strains without constraints imposed by in vitro manipulations. Strains are assembled in a pairwise manner by establishing a donor strain that harbors conjugation machinery and a recipient strain that receives DNA from the donor. Within strain pairs, targeted placement of a conjugal origin of transfer and selectable markers in donor and recipient genomes enables the controlled transfer and selection of desired donor-recipient chimeric genomes. By design, selectable markers act as genomic anchor points, and they are recycled in subsequent rounds of hierarchical genome transfer. A single round of CAGE can be completed in a week, thus enabling four rounds (hierarchical assembly of 16 strains) of CAGE to be completed in roughly 1 month.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Ochman, H., Lawrence, J.G. & Groisman, E.A. Lateral gene transfer and the nature of bacterial innovation. Nature 405, 299–304 (2000).
Smith, G.R. Conjugational recombination in E. coli: myths and mechanisms. Cell 64, 19–27 (1991).
Lederberg, J. & Tatum, E. Gene recombination in Escherichia coli. Nature 53, 673–684 (1946).
Pansegrau, W. et al. Complete nucleotide sequence of Birmingham IncPα plasmids: compilation and comparative analysis. J. Mol. Biol. 239, 623–663 (1994).
Lanka, E. & Wilkins, B.M. DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 64, 141–169 (1995).
Curtiss, R. Bacterial conjugation. Annu. Rev. Microbiol. 23, 69–136 (1969).
Fürste, J. & Pansegrau, W. Conjugative transfer of promiscuous IncP plasmids: interaction of plasmid-encoded products with the transfer origin. Proc. Natl. Acad. Sci. USA 86, 1771–1775 (1989).
Guiney, D. & Yakobson, E. Location and nucleotide sequence of the transfer origin of the broad host range plasmid RK2. Proc. Natl. Acad. Sci. USA 80, 3595–3598 (1983).
Wollman, E.-L., Jacob, F. & Hayes, W. Conjugation and genetic recombination in Escherichia coli K-12. Cold Spring Harb. Symp. Quant. Biol. 21, 141–162 (1956).
Isaacs, F.J. et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science 333, 348–353 (2011).
Court, D.L., Sawitzke, J.A. & Thomason, L.C. Genetic engineering using homologous recombination. Annu. Rev. Genet. 36, 361–388 (2002).
Sharan, S.K., Thomason, L.C., Kuznetsov, S.G. & Court, D.L. Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc. 4, 206–223 (2009).
DeVito, J.A. Recombineering with tolC as a selectable/counter-selectable marker: remodeling the rRNA operons of Escherichia coli. Nucleic Acids Res. 36, e4 (2008).
Schwartz, S.A. & Helinski, D.R. Purification and characterization of colicin E1. J. Biol. Chem. 246, 6318–6327 (1971).
Gallagher, R.R., Li, Z., Lewis, A.O. & Isaacs, F.J. 10.1038/nprot.2014.082. Nat. Protoc. 9, 2301–2316 (2014).
Lajoie, M.J. et al. Genomically recoded organisms expand biological functions. Science 342, 357–360 (2013).
Bates, S., Cashmore, A.M. & Wilkins, B.M. IncP plasmids are unusually effective in mediating conjugation of Escherichia coli and Saccharomyces cerevisiae: involvement of the Tra2 mating system. J. Bacteriol. 180, 6538–6543 (1998).
Thomas, C.M. & Smith, C.A. Incompatibility group P plasmids: genetics, evolution, and use in genetic manipulation. Annu. Rev. Microbiol. 41, 77–101 (1987).
Waters, V.L. Conjugation between bacterial and mammalian cells. Nat. Genet. 231, 375–376 (2001).
Gregg, C.J. et al. Rational optimization of tolC as a powerful dual selectable marker for genome engineering. Nucleic Acids Res. 42, 4779–4790 (2014).
Smith, C., Econome, J. & Schutt, A. A physical map of the Escherichia coli K 12 genome. Science 236, 1448–1453 (1987).
Warming, S., Costantino, N., Court, D.L., Jenkins, N.A. & Copeland, N.G. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33, e36 (2005).
Wong, Q.N.Y. et al. Efficient and seamless DNA recombineering using a thymidylate synthase A selection system in Escherichia coli. Nucleic Acids Res. 33, e59 (2005).
SantaLucia, J. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. USA 95, 1460–1465 (1998).
Acknowledgements
The authors acknowledge support from the U.S. Department of Energy (DE-FG02-02ER63445), Defense Advanced Research Projects Agency (N66001-12-C-4020, N66001-12-C-4211), Arnold and Mabel Beckman Foundation (F.J.I.), Gruber Foundation (N.J.M.), the National Institute of Health Cellular and Molecular Biology Training Grant (N.J.M.), the National Institute of Health Genetics Training Grant (N.J.M.), and the National Institute of Health Biophysics Training Grant (D.W.M.) and the Yale Chemical Biology Institute Training Grant (D.W.M.).
Author information
Authors and Affiliations
Contributions
F.J.I. developed the CAGE methodology in collaboration with A. Tolonen, M. Lajoie and G. Church. N.J.M. finalized the CAGE protocol, wrote the manuscript and created figures. D.W.M. provided feedback on manuscript and created figures.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Table 1
Safe Insertion Regions (SIRs) in E. coli MG1655. (PDF 167 kb)
Supplementary Data
CAGE universal cassettes. (PDF 132 kb)
Rights and permissions
About this article
Cite this article
Ma, N., Moonan, D. & Isaacs, F. Precise manipulation of bacterial chromosomes by conjugative assembly genome engineering. Nat Protoc 9, 2285–2300 (2014). https://doi.org/10.1038/nprot.2014.081
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2014.081
This article is cited by
-
Continuous synthesis of E. coli genome sections and Mb-scale human DNA assembly
Nature (2023)
-
Bioprospecting of unexplored halophilic actinobacteria against human infectious pathogens
3 Biotech (2023)
-
A synthetic ‘essentialome’ for axenic culturing of ‘Candidatus Liberibacter asiaticus’
BMC Research Notes (2022)
-
Building genomes to understand biology
Nature Communications (2020)
-
Total synthesis of Escherichia coli with a recoded genome
Nature (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.