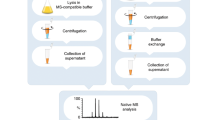
Abstract
The molecular complexity of biopharmaceuticals puts severe demands on the bioanalytical techniques required for their comprehensive structural characterization. Mass spectrometry (MS) has gained importance in the analysis of biopharmaceuticals, taking different complementary approaches ranging from peptide-based sequencing to direct analysis of intact proteins and protein assemblies. In this protocol, we describe procedures optimized to perform the analysis of monoclonal antibodies (mAbs) at the intact protein level under pseudo-native conditions, using native MS. Some of the strengths of native MS in the analysis of biopharmaceuticals are its analysis speed, sensitivity and specificity: for most experiments, the whole protocol requires one working day, whereby tens of samples can be analyzed in a multiplexed manner, making it suitable for high-throughput analysis. This method can be used for different applications such as the analysis of mixtures of mAbs, drug-antibody conjugates and the analysis of mAb post-translational modifications, including the qualitative and quantitative analysis of mAb glycosylation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Reichert, J.M. Marketed therapeutic antibodies compendium. MAbs 4, 413–415 (2012).
Harris, L.J., Skaletsky, E. & McPherson, A. Crystallographic structure of an intact IgG1 monoclonal antibody. J. Mol. Biol. 275, 861–872 (1998).
Saphire, E.O. et al. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science 293, 1155–1159 (2001).
Liu, H., Gaza-Bulseco, G., Faldu, D., Chumsae, C. & Sun, J. Heterogeneity of monoclonal antibodies. J. Pharm. Sci. 97, 2426–2447 (2008).
Jefferis, R. Glycosylation of recombinant antibody therapeutics. Biotechnol. Prog. 21, 11–16 (2005).
Beck, A. et al. Trends in glycosylation, glycoanalysis and glycoengineering of therapeutic antibodies and Fc-fusion proteins. Curr. Pharm. Biotechnol. 9, 482–501 (2008).
Huhn, C., Selman, M.H., Ruhaak, L.R., Deelder, A.M. & Wuhrer, M. IgG glycosylation analysis. Proteomics 9, 882–913 (2009).
Flynn, G.C., Chen, X., Liu, Y.D., Shah, B. & Zhang, Z. Naturally occurring glycan forms of human immunoglobulins G1 and G2. Mol. Immunol. 47, 2074–2082 (2010).
Brady, L.J., Martinez, T. & Balland, A. Characterization of nonenzymatic glycation on a monoclonal antibody. Anal. Chem. 79, 9403–9413 (2007).
Diepold, K. et al. Simultaneous assessment of Asp isomerization and Asn deamidation in recombinant antibodies by LC-MS following incubation at elevated temperatures. PLoS ONE 7, e30295 (2012).
Liu, H. & May, K. Disulfide bond structures of IgG molecules: structural variations, chemical modifications and possible impacts to stability and biological function. MAbs 4, 17–23 (2012).
Li, X. et al. Disulfide bond assignment of an IgG1 monoclonal antibody by LC-MS with post-column partial reduction. Anal. Biochem. 436, 93–100 (2013).
Pace, A.L., Wong, R.L., Zhang, Y.T., Kao, Y.H. & Wang, Y.J. Asparagine deamidation dependence on buffer type, pH, and temperature. J. Pharm. Sci. 102, 1712–1723 (2013).
Logtenberg, T. Antibody cocktails: next-generation biopharmaceuticals with improved potency. Trends Biotechnol. 25, 390–394 (2007).
Robak, T. The emerging therapeutic role of antibody mixtures. Expert Opin. Biol. Ther. 13, 953–958 (2013).
Pro, B. & Perini, G.F. Brentuximab vedotin in Hodgkin′s lymphoma. Expert Opin. Biol. Ther. 12, 1415–1421 (2012).
Sievers, E.L. & Senter, P.D. Antibody-drug conjugates in cancer therapy. Annu. Rev. Med. 64, 15–29 (2013).
Beck, A., Sanglier-Cianferani, S. & Van Dorsselaer, A. Biosimilar, biobetter, and next generation antibody characterization by mass spectrometry. Anal. Chem. 84, 4637–4646 (2012).
Beck, A., Wagner-Rousset, E., Ayoub, D., Van Dorsselaer, A. & Sanglier-Cianferani, S. Characterization of therapeutic antibodies and related products. Anal. Chem. 85, 715–736 (2013).
Gahoual, R. et al. Rapid and multi-level characterization of trastuzumab using sheathless capillary electrophoresis-tandem mass spectrometry. MAbs 5, 479–490 (2013).
Kang, X., Kutzko, J.P., Hayes, M.L. & Frey, D.D. Monoclonal antibody heterogeneity analysis and deamidation monitoring with high-performance cation-exchange chromatofocusing using simple, two component buffer systems. J. Chromatogr. A 1283, 89–97 (2013).
Zhang, Z., Pan, H. & Chen, X. Mass spectrometry for structural characterization of therapeutic antibodies. Mass Spectrom. Rev. 28, 147–176 (2009).
Sharon, M. & Robinson, C.V. The role of mass spectrometry in structure elucidation of dynamic protein complexes. Annu. Rev. Biochem. 76, 167–193 (2007).
Heck, A.J. Native mass spectrometry: a bridge between interactomics and structural biology. Nat. Methods 5, 927–933 (2008).
Konijnenberg, A., Butterer, A. & Sobott, F. Native ion mobility-mass spectrometry and related methods in structural biology. Biochim. Biophys. Acta 1834, 1239–1256 (2013).
Valliere-Douglass, J.F., McFee, W.A. & Salas-Solano, O. Native intact mass determination of antibodies conjugated with monomethyl Auristatin E and F at interchain cysteine residues. Anal. Chem. 84, 2843–2849 (2012).
Thompson, N.J., Rosati, S., Rose, R.J. & Heck, A.J. The impact of mass spectrometry on the study of intact antibodies: from post-translational modifications to structural analysis. Chem. Commun. (Camb) 49, 538–548 (2013).
Zhang, H., Cui, W. & Gross, M.L. Mass spectrometry for the biophysical characterization of therapeutic monoclonal antibodies. FEBS Lett. (2014); 588, 308–317.
Rosati, S. et al. Exploring an Orbitrap analyzer for the characterization of intact antibodies by native mass spectrometry. Angew. Chem. Int. Ed. Engl. 51, 12992–12996 (2012).
Rose, R.J., Damoc, E., Denisov, E., Makarov, A. & Heck, A.J. High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies. Nat Methods 9, 1084–1086 (2012).
Rosati, S. et al. Qualitative and semiquantitative analysis of composite mixtures of antibodies by native mass spectrometry. Anal. Chem. 84, 7227–7232 (2012).
Beck, A. & Reichert, J.M. Therapeutic Fc-fusion proteins and peptides as successful alternatives to antibodies. MAbs 3, 415–416 (2011).
Lynaugh, H., Li, H. & Gong, B. Rapid Fc glycosylation analysis of Fc fusions with IdeS and liquid chromatography mass spectrometry. MAbs 5, 641–645 (2013).
Peters, R.T. et al. Biochemical and functional characterization of a recombinant monomeric factor VIII-Fc fusion protein. J. Thromb. Haemost. 11, 132–141 (2013).
Chelius, D. et al. Structural and functional characterization of the trifunctional antibody catumaxomab. MAbs 2, 309–319 (2010).
Rose, R.J. et al. Quantitative analysis of the interaction strength and dynamics of human IgG4 half molecules by native mass spectrometry. Structure 19, 1274–1282 (2011).
Kukrer, B. et al. Mass spectrometric analysis of intact human monoclonal antibody aggregates fractionated by size-exclusion chromatography. Pharm. Res. 27, 2197–2204 (2010).
Atmanene, C. et al. Extending mass spectrometry contribution to therapeutic monoclonal antibody lead optimization: characterization of immune complexes using noncovalent ESI-MS. Anal. Chem. 81, 6364–6373 (2009).
Rosati, S. et al. In-depth qualitative and quantitative analysis of composite glycosylation profiles and other micro-heterogeneity on intact monoclonal antibodies by high-resolution native mass spectrometry using a modified Orbitrap. MAbs 5, 917–924 (2013).
Shi, Y., Li, Z., Qiao, Y. & Lin, J. Development and validation of a rapid capillary zone electrophoresis method for determining charge variants of mAb. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 906, 63–68 (2012).
Xu, W. et al. Method to convert N-terminal glutamine to pyroglutamate for characterization of recombinant monoclonal antibodies. Anal. Biochem. 436, 10–12 (2013).
Thompson, N.J., Hendriks, L.J.A., de Kruif, J., Throsby, M. & Heck, A.J.R. Complex mixtures of antibodies generated from a single production qualitatively and quantitatively evaluated by native Orbitrap mass spectrometry. MAbs 6, 197–203 (2014).
van den Heuvel, R.H. et al. Improving the performance of a quadrupole time-of-flight instrument for macromolecular mass spectrometry. Anal. Chem. 78, 7473–7483 (2006).
Sobott, F., Hernandez, H., McCammon, M.G., Tito, M.A. & Robinson, C.V. A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal. Chem. 74, 1402–1407 (2002).
Fenn, J.B., Mann, M., Meng, C.K., Wong, S.F. & Whitehouse, C.M. Electrospray ionization for mass spectrometry of large biomolecules. Science 246, 64–71 (1989).
Wilm, M. & Mann, M. Analytical properties of the nanoelectrospray ion source. Anal. Chem. 68, 1–8 (1996).
Almeida, R. et al. Coupling of fully automated chip-based electrospray ionization to high-capacity ion trap mass spectrometer for ganglioside analysis. Anal. Biochem. 378, 43–52 (2008).
Zhang, S., Van Pelt, C.K. & Henion, J.D. Automated chip-based nanoelectrospray-mass spectrometry for rapid identification of proteins separated by two-dimensional gel electrophoresis. Electrophoresis 24, 3620–3632 (2003).
Hernandez, H. & Robinson, C.V. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat. Protoc. 2, 715–726 (2007).
Acknowledgements
This work, and in particular that of S.R., A.B. and A.J.R.H., was supported in part by Stichting voor de Technische Wetenschappen (STW) (project 10805) and Y.Y. and A.J.R.H. are supported by the ManiFold project, grant agreement number 317371. We further acknowledge support of the PRIME-XS project, grant agreement number 262067, funded by the European Union Seventh Framework Program. The Netherlands Proteomics Centre, embedded in The Netherlands Genomics Initiative, is acknowledged for funding.
Author information
Authors and Affiliations
Contributions
S.R., Y.Y., A.B. and A.J.R.H. developed the protocols described here, obtained the presented data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Rosati, S., Yang, Y., Barendregt, A. et al. Detailed mass analysis of structural heterogeneity in monoclonal antibodies using native mass spectrometry. Nat Protoc 9, 967–976 (2014). https://doi.org/10.1038/nprot.2014.057
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2014.057
This article is cited by
-
Standard Proteoforms and Their Complexes for Native Mass Spectrometry
Journal of the American Society for Mass Spectrometry (2019)
-
Distinct Stabilities of the Structurally Homologous Heptameric Co-Chaperonins GroES and gp31
Journal of the American Society for Mass Spectrometry (2019)
-
A rapid and quantitative technique for assessing IgG monomeric purity, calibrated with the NISTmAb reference material
Analytical and Bioanalytical Chemistry (2019)
-
Native mass spectrometry combined with enzymatic dissection unravels glycoform heterogeneity of biopharmaceuticals
Nature Communications (2018)
-
Direct quality control of glycoengineered erythropoietin variants
Nature Communications (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.