Abstract
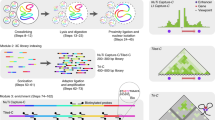
Chromosome conformation capture (3C) is a powerful technique for analyzing spatial chromatin organization in vivo. Technical variants of the assay ('4C') allow the systematic detection of genome-wide coassociations with bait sequences of interest, enabling the nuclear environments of specific genes to be probed. We describe enhanced 4C (e4C, enhanced chromosome conformation capture on chip), a technique incorporating additional enrichment steps for bait-specific sequences, and thus improving sensitivity in the detection of weaker, distal chromatin coassociations. In brief, e4C entails the fixation, restriction digestion and ligation steps of conventional 3C, with an optional chromatin immunoprecipitation (ChIP) step to select for subsets of chromatin coassociations, followed by bait enrichment by biotinylated primer extension and pull-down, adapter ligation and PCR amplification. Chromatin coassociations with the bait sequence can then be assessed by hybridizing e4C products to microarrays or sequencing. The e4C procedure takes approximately 1 week to go from tissue to DNA ready for microarray hybridization.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Schoenfelder, S. et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat. Genet. 42, 53–61 (2010).
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002).
Hagege, H. et al. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2, 1722–1733 (2007).
Sexton, T., Bantignies, F. & Cavalli, G. Genomic interactions: chromatin loops and gene meeting points in transcriptional regulation. Semin. Cell Dev. Biol. 20, 849–855 (2009).
Lomvardas, S. et al. Interchromosomal interactions and olfactory receptor choice. Cell 126, 403–413 (2006).
Simonis, M. et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 38, 1348–1354 (2006).
Wurtele, H. & Chartrand, P. Genome-wide scanning of HoxB1-associated loci in mouse ES cells using an open-ended chromosome conformation capture methodology. Chromosome Res. 14, 477–495 (2006).
Zhao, Z. et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 38, 1341–1347 (2006).
Ohlsson, R. & Gondor, A. The 4C technique: the 'Rosetta stone' for genome biology in 3D? Curr. Opin. Cell Biol. 19, 321–325 (2007).
Hakim, O. et al. Diverse gene reprogramming events occur in the same spatial clusters of distal regulatory elements. Genome Res. 21, 697–706 (2011).
Noordermeer, D. et al. Variegated gene expression caused by cell-specific long-range DNA interactions. Nat. Cell Biol. 13, 944–951 (2011).
Dostie, J. et al. Chromosome conformation capture carbon copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 16, 1299–1309 (2006).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Fullwood, M.J. et al. An oestrogen receptor-α–bound human chromatin interactome. Nature 462, 58–64 (2009).
Horike, S., Cai, S., Miyano, M., Cheng, J.F. & Kohwi-Shigematsu, T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat. Genet. 37, 31–40 (2005).
Louwers, M., Splinter, E., van Driel, R., de Laat, W. & Stam, M. Studying physical chromatin interactions in plants using chromosome conformation capture (3C). Nat. Protoc. 4, 1216–1229 (2009).
Raab, J.R. et al. Human tRNA genes function as chromatin insulators. EMBO J. (2011); 31, 330–350.
Brazma, A. et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 29, 365–371 (2001).
de Wit, E., Braunschweig, U., Greil, F., Bussemaker, H.J. & van Steensel, B. Global chromatin domain organization of the Drosophila genome. PLoS Genet. 4, e1000045 (2008).
Bantignies, F. et al. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell 144, 214–226 (2011).
Tolhuis, B. et al. Interactions among polycomb domains are guided by chromosome architecture. PLoS Genet. 7, e1001343 (2011).
Kilkenny, C., Browne, W.J., Cuthill, I.C., Emerson, M. & Altman, D.G. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 8, e1000412 (2010).
Comet, I., Schuettengruber, B., Sexton, T. & Cavalli, G. A chromatin insulator driving three-dimensional Polycomb response element (PRE) contacts and Polycomb association with the chromatin fiber. Proc. Natl. Acad. Sci. USA 108, 2294–2299 (2011).
Sandmann, T., Jakobsen, J.S. & Furlong, E.E. ChIP-on-chip protocol for genome-wide analysis of transcription factor binding in Drosophila melanogaster embryos. Nat. Protoc. 1, 2839–2855 (2006).
Osborne, C.S. et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 36, 1065–1071 (2004).
Carter, D., Chakalova, L., Osborne, C.S., Dai, Y.F. & Fraser, P. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 32, 623–626 (2002).
Tolhuis, B., Palstra, R.J., Splinter, E., Grosveld, F. & de Laat, W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell 10, 1453–1465 (2002).
Acknowledgements
This work was supported by the UK Medical Research Council, the UK Biotechnology and Biological Sciences Research Council and by a long-term EMBO fellowship to D.U.
Author information
Authors and Affiliations
Contributions
T.S., S.K. and P.F. designed the experiments, T.S. and S.K. developed the method, J.A.M. and D.U. optimized the chromatin immunoprecipitation steps, T.N. developed primers for restriction digestion efficiency tests and T.S. and P.F. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Typical raw e4C and ChIP-e4C results, using Hbb-b1 sequence as bait. (PDF 360 kb)
Supplementary Table 1
List of primers used for assessing BglII restriction digestion efficiency in mouse tissues. (PDF 72 kb)
Supplementary Table 2
List of 3C primers used in Anticipated Results section. (PDF 52 kb)
Supplementary Table 3
List of ChIP primers used in Anticipated Results section. (PDF 52 kb)
Supplementary Table 4
List of primers used for assessing e4C bait enrichment in Anticipated Results section. (PDF 51 kb)
Rights and permissions
About this article
Cite this article
Sexton, T., Kurukuti, S., Mitchell, J. et al. Sensitive detection of chromatin coassociations using enhanced chromosome conformation capture on chip. Nat Protoc 7, 1335–1350 (2012). https://doi.org/10.1038/nprot.2012.071
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2012.071
This article is cited by
-
Hippocampal Cannabinoid 1 Receptors Are Modulated Following Cocaine Self-administration in Male Rats
Molecular Neurobiology (2022)
-
Epigenetic specifications of host chromosome docking sites for latent Epstein-Barr virus
Nature Communications (2020)
-
CTCF-mediated genome organization and leukemogenesis
Leukemia (2020)
-
Novel insights into chromosomal conformations in cancer
Molecular Cancer (2017)
-
Chromatin states and nuclear organization in development — a view from the nuclear lamina
Genome Biology (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.