Abstract
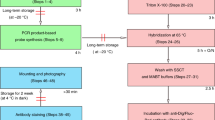
In situ hybridization (ISH) is widely used to study the spatial distribution of gene expression in developing embryos. It is the method of choice to analyze the normal pattern of expression of a gene and also to characterize how the expression of a gene, or a group of genes, is altered in response to experimental or genetic manipulations. The standard protocols for this technique use a chromogenic reaction that produces a purple or red precipitate in cells expressing the target gene. This technique has significant disadvantages when compared with fluorescent techniques, as it cannot detect regions of overlap and external staining masks internal staining. We present a protocol for three-channel fluorescent ISH (FISH) optimized for wholemount analysis of large vertebrate embryos. Multichannel FISH in combination with immunofluorescence or chromogenic ISH offers a suite of approaches that allow accurate mapping of overlapping gene expression patterns in two- and three-dimensions. The time required for the protocol varies depending on the number of channels sampled and ranges from 3 to 5 d plus an additional 2 d to completely wash embryos and prepare for documentation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
McDougall, J.K., Dunn, A.R. & Jones, K.W. In situ hybridization of adenovirus RNA and DNA. Nature 236, 346–348 (1972).
Melton, D.A. Translocation of a localized maternal mRNA to the vegetal pole of Xenopus oocytes. Nature 328, 80–82 (1987).
Wilkinson, D.G., Bhatt, S., Chavrier, P., Bravo, R. & Charnay, P. Segment-specific expression of a zinc-finger gene in the developing nervous system of the mouse. Nature 337, 461–464 (1989).
Tautz, D. & Pfeifle, C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98, 81–85 (1989).
Sambrook, J. & Russell, D.W. Molecular Cloning, A Laboratory Manual 3rd edn. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2001).
Brivanlou, A.H. & Harland, R.M. Expression of an engrailed-related protein is induced in the anterior neural ectoderm of early Xenopus embryos. Development 106, 611–617 (1989).
Harland, R.M. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 36, 685–695 (1991).
Thisse, C. & Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 (2008).
Acloque, H., Wilkinson, D.G. & Nieto, M.A. Chapter 9 in situ hybridization analysis of chick embryos in whole-mount and tissue sections. Methods Cell Biol. 87, 169–185 (2008).
Hargrave, M., Bowles, J. & Koopman, P. In situ Hybridization Protocols. In Methods in Molecular Biology (eds. Darby, I.A. & Hewitson, T.D.) 103–114 (Humana, Totowa, New Jersey, 2006).
Pollet, N. et al. An atlas of differential gene expression during early Xenopus embryogenesis. Mech. Dev. 122, 365–439 (2005).
Magdaleno, S. et al. BGEM: an in situ hybridization database of gene expression in the embryonic and adult mouse nervous system. PLoS Biol. 4, e86 (2006).
Bell, G.W., Yatskievych, T.A. & Antin, P.B. GEISHA, a whole-mount in situ hybridization gene expression screen in chicken embryos. Dev. Dyn. 229, 677–687 (2004).
Hauptmann, G. One-, two-, and three-color whole-mount in situ hybridization to Drosophila embryos. Methods 23, 359–372 (2001).
Bueno, D., Skinner, J., Abud, H. & Heath, J.K. Double in situ hybridization on mouse embryos for detection of overlapping regions of gene expression. Trends Genet. 12, 385–387 (1996).
Davidson, L.A. & Keller, R.E. Neural tube closure in Xenopus laevis involves medial migration, directed protrusive activity, cell intercalation and convergent extension. Development 126, 4547–4556 (1999).
Zhou, X. & Vize, P.D. Proximo-distal specialization of epithelial transport processes within the Xenopus pronephric kidney tubules. Dev. Biol. 271, 322–338 (2004).
Kosman, D. et al. Multiplex detection of RNA expression in Drosophila embryos. Science 305, 846 (2004).
Urban, A.E. et al. FGF is essential for both condensation and mesenchymal–epithelial transition stages of pronephric kidney tubule development. Dev. Biol. 297, 103–117 (2006).
Gerth, V.E., Zhou, X. & Vize, P.D. Nephrin expression and three-dimensional morphogenesis of the Xenopus pronephric glomus. Dev. Dyn. 233, 1131–1139 (2005).
Denkers, N., García-Villalba, P., Rodesch, C.K., Nielson, K.R. & Mauch, T.J. FISHing for chick genes: triple-label whole-mount fluorescence in situ hybridization detects simultaneous and overlapping gene expression in avian embryos. Dev. Dyn. 229, 651–657 (2004).
Vize, P.D., Jones, E.A. & Pfister, R. Development of the Xenopus pronephric system. Dev. Biol. 171, 531–540 (1995).
Acknowledgements
These protocols are adaptations of those originally developed by Richard Harland and Lance Davidson. The Photoshop image manipulation pipeline was developed by Victor Gerth, who also performed the data processing for the 3D models. This work was supported by grants from the AHFMR to P.D.V.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Movie 1: Two channel FISH sampled in 3D.
Two channel FISH illustrating expression of nephrin in the pronephric glomus and atp1a1 in the pronephric nephron. Sampling by confocal microscopy. (MOV 1334 kb)
Rights and permissions
About this article
Cite this article
Vize, P., McCoy, K. & Zhou, X. Multichannel wholemount fluorescent and fluorescent/chromogenic in situ hybridization in Xenopus embryos. Nat Protoc 4, 975–983 (2009). https://doi.org/10.1038/nprot.2009.69
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2009.69
This article is cited by
-
Combined whole-mount fluorescence in situ hybridization and antibody staining in zebrafish embryos and larvae
Nature Protocols (2020)
-
Toolbox in a tadpole: Xenopus for kidney research
Cell and Tissue Research (2017)
-
Yap-dependent reprogramming of Lgr5+ stem cells drives intestinal regeneration and cancer
Nature (2015)
-
Optimized RNA ISH, RNA FISH and protein-RNA double labeling (IF/FISH) in Drosophila ovaries
Nature Protocols (2013)
-
Fluorescent mRNA labeling through cytoplasmic FISH
Nature Protocols (2013)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.