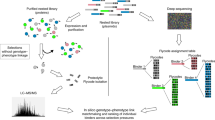
Abstract
Signaling complexes usually involve multidomain proteins containing catalytic domains and peptide recognition modules (PRMs), which mediate protein–protein interactions and assemble complexes by binding to ligands containing a core sequence motif. Concomitant to large-scale physical interaction screening, considerable effort has been devoted toward the elucidation of consensus profiles for common PRMs. We describe herein a robust and proven protocol to generate consensus profiles for PRMs using phage-displayed peptide libraries. The initial phase of the protocol entails the cloning, expression and purification of PRMs as fusion proteins, in addition to the construction of highly diverse phage-displayed peptide libraries. The affinity selection process described thereafter enables a single researcher to efficiently probe the recognition profiles of numerous PRMs in a 1 week time period.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Cesareni, G. Modular Protein Domains, xxii, 501 p. (Wiley-VCH, Weinheim, 2005).
Letunic, I. et al. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 34, D257–D260 (2006).
Pawson, T. & Nash, P. Assembly of cell regulatory systems through protein interaction domains. Science 300, 445–452 (2003).
Harris, B.Z. & Lim, W.A. Mechanism and role of PDZ domains in signaling complex assembly. J. Cell Sci. 114, 3219–3231 (2001).
Zarrinpar, A., Bhattacharyya, R.P. & Lim, W.A. The structure and function of proline recognition domains. Sci STKE 2003, RE8 (2003).
Tong, A.H. et al. A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science 295, 321–324 (2002).
Uetz, P. et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae . Nature 403, 623–627 (2000).
Ito, T. et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98, 4569–4574 (2001).
Li, S. et al. A map of the interactome network of the metazoan C. elegans . Science 303, 540–543 (2004).
Giot, L. et al. A protein interaction map of Drosophila melanogaster . Science 302, 1727–1736 (2003).
Rual, J.F. et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature 437, 1173–1178 (2005).
Stelzl, U. et al. A human protein–protein interaction network: a resource for annotating the proteome. Cell 122, 957–968 (2005).
Yang, M., Wu, Z. & Fields, S. Protein-peptide interactions analyzed with the yeast two-hybrid system. Nucleic Acids Res. 23, 1152–1156 (1995).
Song, E. et al. A high efficiency strategy for binding property characterization of peptide-binding domains. Mol. Cell. Proteomics 5, 1368–1381 (2006).
Landgraf, C. et al. Protein interaction networks by proteome peptide scanning. PLoS Biol. 2, E14 (2004).
Smith, M.J., Hardy, W.R., Murphy, J.M., Jones, N. & Pawson, T. Screening for PTB domain binding partners and ligand specificity using proteome-derived NPXY peptide arrays. Mol. Cell. Biol. 26, 8461–8474 (2006).
Sidhu, S.S. Phage display in pharmaceutical biotechnology. Curr. Opin. Biotechnol. 11, 610–616 (2000).
Smith, G.P. & Petrenko, V.A. Phage display. Chem. Rev. 97, 391–410 (1997).
Georgiou, G. et al. Display of heterologous proteins on the surface of microorganisms: from the screening of combinatorial libraries to live recombinant vaccines. Nat. Biotechnol. 15, 29–34 (1997).
Wittrup, K.D. Protein engineering by cell-surface display. Curr. Opin. Biotechnol. 12, 395–399 (2001).
Cull, M., Miller, J. & Schatz, P. Screening for receptor ligands using libraries of peptides linked to the C terminus of the lac repressor. Proc. Natl. Acad. Sci. USA 89, 1865–1869 (1992).
Lipovsek, D. & Pluckthun, A. In-vitro protein evolution by ribosome display and mRNA display. J. Immunol. Methods 290, 51–67 (2004).
Sidhu, S.S., Lowman, H.B., Cunningham, B.C. & Wells, J.A. Phage display &1QJ;for selection of novel binding peptides. Methods Enzymol. 328, 333–363 (2000).
Sidhu, S.S. Phage display: increasing the rewards from genomic information. Drug Discov. Today 6, 936 (2001).
Feng, S., Kasahara, C., Rickles, R.J. & Schreiber, S.L. Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands. Proc. Natl. Acad. Sci. USA 92, 12408–12415 (1995).
Rickles, R.J. et al. Phage display selection of ligand residues important for Src homology 3 domain binding specificity. Proc. Natl. Acad. Sci. USA 92, 10909–10913 (1995).
Sparks, A.B. et al. Distinct ligand preferences of Src homology 3 domains from Src, Yes, Abl, cortactin, p53bp2, PLCgamma, Crk, and Grb2. Proc. Natl. Acad. Sci. USA 93, 1540–1544 (1996).
Kasanov, J., Pirozzi, G., Uveges, A.J. & Kay, B.K. Characterizing class I WW domains defines key specificity determinants and generates mutant domains with novel specificities. Chem. Biol. 8, 231–241 (2001).
Zhang, Y. et al. Convergent and divergent ligand specificity among PDZ domains of the LAP and zonula occludens (ZO) families. J. Biol. Chem. 281, 22299–22311 (2006).
Appleton, B.A. et al. Comparative structural analysis of the Erbin PDZ domain and the first PDZ domain of ZO-1. Insights into determinants of PDZ domain specificity. J. Biol. Chem. 281, 22312–22320 (2006).
Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Bateman, A. et al. The Pfam protein families database. Nucleic Acids Res. 32, D138–D141 (2004).
Aslanidis, C. & de Jong, P.J. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 18, 6069–6074 (1990).
Petrenko, V.A. & Smith, G.P. Vectors and modes of display. in Phage Display in Biotechnology and Drug Discovery Vol. 3 (ed. Sidhu, S.S.) 63–110 (Taylor and Francis Group, Boca Raton, FL, 2005).
Fellouse, F.A. & Pal, G. Methods for the construction of phage-displayed libraries. in Phage Display in Biotechnology and Drug Discovery Vol. 3 (ed. Sidhu, S.S.) 111–142 (Taylor and Francis Group, Boca Raton, FL, 2005).
Sidhu, S.S., Feld, B.K. & Weiss, G.A. M13 bacteriophage coat proteins engineered for improved phage display. Methods Mol. Biol. 352, 205–219 (2007).
Kunkel, T.A., Roberts, J.D. & Zakour, R.A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154, 367–382 (1987).
Laura, R.P. et al. The Erbin PDZ domain binds with high affinity and specificity to the carboxyl termini of delta-catenin and ARVCF. J. Biol. Chem. 277, 12906–12914 (2002).
Higgins, D.G., Thompson, J.D. & Gibson, T.J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266, 383–402 (1996).
Crooks, G.E., Hon, G., Chandonia, J.M. & Brenner, S.E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Maniatis, T., Fritsch, E.F. & Sambrook, J. Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, 1987).
Lechner, R.L., Engler, M.J. & Richardson, C.C. Characterization of strand displacement synthesis catalyzed by bacteriophage T7 DNA polymerase. J. Biol. Chem. 258, 11174–11184 (1983).
Held, H.A. & Sidhu, S.S. Comprehensive mutational analysis of the M13 major coat protein: improved scaffolds for C-terminal phage display. J. Mol. Biol. 340, 587–597 (2004).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Tonikian, R., Zhang, Y., Boone, C. et al. Identifying specificity profiles for peptide recognition modules from phage-displayed peptide libraries. Nat Protoc 2, 1368–1386 (2007). https://doi.org/10.1038/nprot.2007.151
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2007.151
This article is cited by
-
CD98hc is a target for brain delivery of biotherapeutics
Nature Communications (2023)
-
A potent synthetic nanobody with broad-spectrum activity neutralizes SARS-CoV-2 virus and the Omicron variant BA.1 through a unique binding mode
Journal of Nanobiotechnology (2022)
-
Engineering an autonomous VH domain to modulate intracellular pathways and to interrogate the eIF4F complex
Nature Communications (2022)
-
convertibleCARs: A chimeric antigen receptor system for flexible control of activity and antigen targeting
Communications Biology (2020)
-
A protein scaffold, engineered SPINK2, for generation of inhibitors with high affinity and specificity against target proteases
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.