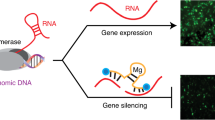
Abstract
RNA interference is a powerful tool for target-specific knockdown of gene expression. The triggers for this process are duplex small interfering RNAs (siRNAs) of 21–25 nt with 2-bp 3′ overhangs produced in cells by the RNase III family member Dicer. We have observed that short RNAs that are long enough to serve as Dicer substrates (D-siRNA) can often evoke more potent RNA interference than the corresponding 21-nt siRNAs; this is probably a consequence of the physical handoff of the Dicer-produced siRNAs to the RNA-induced silencing complex. Here we describe the design parameters for D-siRNAs and a protocol for in vitro and in vivo intraperitoneal delivery of D-siRNAs and siRNAs to macrophages. siRNA delivery and transfection and analysis of macrophages in vivo can be accomplished within 36 h.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Elbashir, S.M. et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498 (2001).
Elbashir, S.M., Lendeckel, W. & Tuschl, T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15, 188–200 (2001).
Khvorova, A., Reynolds, A. & Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 115, 209–216 (2003).
Schwarz, D.S. et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115, 199–208 (2003).
Amarzguioui, M., Holen, T., Babaie, E. & Prydz, H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 31, 589–595 (2003).
Czauderna, F. et al. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 31, 2705–2716 (2003).
Dande, P. et al. Improving RNA interference in mammalian cells by 4′-thio-modified small interfering RNA (siRNA): effect on siRNA activity and nuclease stability when used in combination with 2′-O-alkyl modifications. J. Med. Chem. 49, 1624–1634 (2006).
Morrissey, D.V. et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 23, 1002–1007 (2005).
Prakash, T.P. et al. Positional effect of chemical modifications on short interference RNA activity in mammalian cells. J. Med. Chem. 48, 4247–4253 (2005).
Braasch, D.A. et al. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry 42, 7967–7975 (2003).
Chiu, Y.L. & Rana, T.M. siRNA function in RNAi: a chemical modification analysis. RNA 9, 1034–1048 (2003).
Elmen, J. et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 33, 439–447 (2005).
Kim, D.H. et al. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 23, 222–226 (2005).
Rose, S.D. et al. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 33, 4140–4156 (2005).
Siolas, D. et al. Synthetic shRNAs as potent RNAi triggers. Nat. Biotechnol. 23, 227–231 (2005).
Pham, J.W., Pellino, J.L., Lee, Y.S., Carthew, R.W. & Sontheimer, E.J. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell 117, 83–94 (2004).
Gregory, R.I., Chendrimada, T.P., Cooch, N. & Shiekhattar, R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123, 631–640 (2005).
Maniataki, E. & Mourelatos, Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 19, 2979–2990 (2005).
Krol, J. et al. Structural features of microRNA (miRNA) precursors and their relevance to miRNA biogenesis and small interfering RNA/short hairpin RNA design. J. Biol. Chem. 279, 42230–42239 (2004).
Amarzguioui, M. & Prydz, H. An algorithm for selection of functional siRNA sequences. Biochem. Biophys. Res. Commun. 316, 1050–1058 (2004).
Huesken, D. et al. Design of a genome-wide siRNA library using an artificial neural network. Nat. Biotechnol. 23, 995–1001 (2005).
Reynolds, A. et al. Rational siRNA design for RNA interference. Nat. Biotechnol. 22, 326–330 (2004).
Du, Q., Thonberg, H., Wang, J., Wahlestedt, C. & Liang, Z. A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites. Nucleic Acids Res. 33, 1671–1677 (2005).
Elbashir, S.M., Martinez, J., Patkaniowska, A., Lendeckel, W. & Tuschl, T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 20, 6877–6888 (2001).
Holen, T., Amarzguioui, M., Wiiger, M.T., Babaie, E. & Prydz, H. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res. 30, 1757–1766 (2002).
Miller, V.M., Gouvion, C.M., Davidson, B.L. & Paulson, H.L. Targeting Alzheimer's disease genes with RNA interference: an efficient strategy for silencing mutant alleles. Nucleic Acids Res. 32, 661–668 (2004).
Birmingham, A. et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods 3, 199–204 (2006).
Hornung, V. et al. Sequence-specific potent induction of IFN-α by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 11, 263–270 (2005).
Judge, A.D. et al. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat. Biotechnol. 23, 457–462 (2005).
Kariko, K., Bhuyan, P., Capodici, J. & Weissman, D. Small interfering RNAs mediate sequence-independent gene suppression and induce immune activation by signaling through toll-like receptor 3. J. Immunol. 172, 6545–6549 (2004).
Robbins, M.A. & Rossi, J.J. Sensing the danger in RNA. Nat. Med. 11, 250–251 (2005).
Sioud, M. Induction of inflammatory cytokines and interferon responses by double-stranded and single-stranded siRNAs is sequence-dependent and requires endosomal localization. J. Mol. Biol. 348, 1079–1090 (2005).
Marques, J.T. et al. A structural basis for discriminating between self and nonself double-strand RNAs in mammalian cells. Nat. Biotechnol. 24, 559–565 (2006).
Judge, A.D., Bola, G., Lee, A.C. & MacLachlan, I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol. Ther. 13, 494–505 (2006).
Amarzguioui, M. Improved siRNA-mediated silencing in refractory adherent cell lines by detachment and transfection in suspension. Biotechniques 36, 766–768, 770 (2004).
Acknowledgements
J.J.R. was supported by National Institutes of Health grants AI29329, AI424552 and HL07470. E.C. was supported by National Institutes of Health grant EY013814.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
J.H. is a co-founder and employee of Mirus Bio Corporation.
Rights and permissions
About this article
Cite this article
Amarzguioui, M., Lundberg, P., Cantin, E. et al. Rational design and in vitro and in vivo delivery of Dicer substrate siRNA. Nat Protoc 1, 508–517 (2006). https://doi.org/10.1038/nprot.2006.72
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.72
This article is cited by
-
Drug conjugates for targeting regulatory T cells in the tumor microenvironment: guided missiles for cancer treatment
Experimental & Molecular Medicine (2023)
-
Sequence determinant of small RNA production by DICER
Nature (2023)
-
Synthesis and use of an amphiphilic dendrimer for siRNA delivery into primary immune cells
Nature Protocols (2021)
-
Aptamer-miRNA-212 Conjugate Sensitizes NSCLC Cells to TRAIL
Molecular Therapy - Nucleic Acids (2016)
-
Knockdown of β-catenin with Dicer-Substrate siRNAs Reduces Liver Tumor Burden In vivo
Molecular Therapy (2014)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.