Abstract
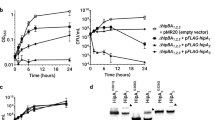
Bacterial toxin–antitoxin systems (TASs) are thought to respond to various stresses, often inducing growth-arrested (persistent) sub-populations of cells whose housekeeping functions are inhibited. Many such TASs induce this effect through the translation-dependent RNA cleavage (RNase) activity of their toxins, which are held in check by their cognate antitoxins in the absence of stress. However, it is not always clear whether specific mRNA targets of orthologous RNase toxins are responsible for their phenotypic effect, which has made it difficult to accurately place the multitude of TASs within cellular and adaptive regulatory networks. Here, we show that the TAS HigBA of Caulobacter crescentus can promote and inhibit bacterial growth dependent on the dosage of HigB, a toxin regulated by the DNA damage (SOS) repressor LexA in addition to its antitoxin HigA, and the target selectivity of HigB's mRNA cleavage activity. HigB reduced the expression of an efflux pump that is toxic to a polarity control mutant, cripples the growth of cells lacking LexA, and targets the cell cycle circuitry. Thus, TASs can have outcome switching activity in bacterial adaptive (stress) and systemic (cell cycle) networks.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Cohen, N. R., Lobritz, M. A. & Collins, J. J. Microbial persistence and the road to drug resistance. Cell Host Microbe 13, 632–642 (2013).
Gerdes, K., Christensen, S. K. & Lobner-Olesen, A. Prokaryotic toxin–antitoxin stress response loci. Nature Rev. Microbiol. 3, 371–382 (2005).
Lewis, K. Persister cells. Annu. Rev. Microbiol. 64, 357–372 (2010).
Schuster, C. F. & Bertram, R. Toxin–antitoxin systems are ubiquitous and versatile modulators of prokaryotic cell fate. FEMS Microbiol. Lett. 340, 73–85 (2013).
Maisonneuve, E., Castro-Camargo, M. & Gerdes, K. (p)ppGpp controls bacterial persistence by stochastic induction of toxin–antitoxin activity. Cell 154, 1140–1150 (2013).
Wang, X. et al. Antitoxin MqsA helps mediate the bacterial general stress response. Nature Chem. Biol. 7, 359–366 (2011).
Maisonneuve, E., Shakespeare, L. J., Jorgensen, M. G. & Gerdes, K. Bacterial persistence by RNA endonucleases. Proc. Natl Acad. Sci. USA 108, 13206–13211 (2011).
Pandey, D. P. & Gerdes, K. Toxin–antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33, 966–976 (2005).
Christensen-Dalsgaard, M. & Gerdes, K. Two higBA loci in the Vibrio cholerae superintegron encode mRNA cleaving enzymes and can stabilize plasmids. Mol. Microbiol. 62, 397–411 (2006).
Christensen-Dalsgaard, M., Jorgensen, M. G. & Gerdes, K. Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol. Microbiol. 75, 333–348 (2010).
Hurley, J. M. & Woychik, N. A. Bacterial toxin HigB associates with ribosomes and mediates translation-dependent mRNA cleavage at A-rich sites. J. Biol. Chem. 284, 18605–18613 (2009).
Schureck, M. A., Dunkle, J. A., Maehigashi, T., Miles, S. J. & Dunham, C. M. Defining the mRNA recognition signature of a bacterial toxin protein. Proc. Natl Acad. Sci. USA 112, 13862–13867 (2015).
Schuessler, D. L. et al. Induced ectopic expression of HigB toxin in Mycobacterium tuberculosis results in growth inhibition, reduced abundance of a subset of mRNAs and cleavage of tmRNA. Mol. Microbiol. 90, 195–207 (2013).
Fiebig, A., Rojas, C. M., Siegal-Gaskins, D. & Crosson, S. Interaction specificity, toxicity, and regulation of a paralogous set of ParE/RelE-family toxin–antitoxin systems. Mol. Microbiol. 77, 236–251 (2010).
Huitema, E., Pritchard, S., Matteson, D., Radhakrishnan, S. K. & Viollier, P. H. Bacterial birth scar proteins mark future flagellum assembly site. Cell 124, 1025–1037 (2006).
Lam, H., Schofield, W. B. & Jacobs-Wagner, C. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell 124, 1011–1023 (2006).
Schofield, W. B., Lim, H. C. & Jacobs-Wagner, C. Cell cycle coordination and regulation of bacterial chromosome segregation dynamics by polarly localized proteins. EMBO J. 29, 3068–3081 (2010).
Kirkpatrick, C. L. & Viollier, P. H. Synthetic interaction between the TipN polarity factor and an AcrAB-family efflux pump implicates cell polarity in bacterial drug resistance. Chem. Biol. 21, 657–665 (2014).
Murray, S. M., Panis, G., Fumeaux, C., Viollier, P. H. & Howard, M. Computational and genetic reduction of a cell cycle to its simplest, primordial components. PLoS Biol. 11, e1001749 (2013).
Da Rocha, R. P., Paquola, A. C., Marques Mdo, V., Menck, C. F. & Galhardo, R. S. Characterization of the SOS regulon of Caulobacter crescentus. J. Bacteriol. 190, 1209–1218 (2008).
Modell, J. W., Kambara, T. K., Perchuk, B. S. & Laub, M. T. A DNA damage-induced, SOS-independent checkpoint regulates cell division in Caulobacter crescentus. PLoS Biol. 12, e1001977 (2014).
Linder, P., Lemeille, S. & Redder, P. Transcriptome-wide analyses of 5′-ends in RNase J mutants of a gram-positive pathogen reveal a role in RNA maturation, regulation and degradation. PLoS Genet. 10, e1004207 (2014).
Redder, P . in RNA Remodeling Proteins Vol. 1259 (ed. Boudvillain, M. ) 69–85 (2015).
Neubauer, C. et al. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell 139, 1084–1095 (2009).
Domian, I. J., Quon, K. C. & Shapiro, L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell 90, 415–424 (1997).
Pedersen, K. et al. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112, 131–140 (2003).
Zhang, J. H., Chung, T. D. & Oldenburg, K. R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen 4, 67–73 (1999).
Heeb, S. et al. Quinolones: from antibiotics to autoinducers. FEMS Microbiol. Rev. 35, 247–274 (2010).
Engelberg-Kulka, H., Amitai, S., Kolodkin-Gal, I. & Hazan, R. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2, e135 (2006).
Ely, B. Genetics of Caulobacter crescentus. Methods Enzymol. 204, 372–384 (1991).
Viollier, P. H. et al. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc. Natl Acad. Sci. USA 101, 9257–9262 (2004).
Marks, M. E. et al. The genetic basis of laboratory adaptation in Caulobacter crescentus. J. Bacteriol. 192, 3678–3688 (2010).
Miller, J. H. in Experiments in Molecular Genetics (ed. Miller, J. H. ) 352–355 (Cold Spring Harbor Laboratory Press, 1972).
Radhakrishnan, S. K., Thanbichler, M. & Viollier, P. H. The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus. Genes Dev. 22, 212–225 (2008).
Fumeaux, C. et al. Cell cycle transition from S-phase to G1 in Caulobacter is mediated by ancestral virulence regulators. Nature Commun. 5, 4081 (2014).
Radhakrishnan, S. K., Pritchard, S. & Viollier, P. H. Coupling prokaryotic cell fate and division control with a bifunctional and oscillating oxidoreductase homolog. Dev. Cell 18, 90–101 (2010).
White, C. L., Kitich, A. & Gober, J. W. Positioning cell wall synthetic complexes by the bacterial morphogenetic proteins MreB and MreD. Mol. Microbiol. 76, 616–633 (2010).
Yasrebi, H. & Redder, P. EMOTE-conv: a computational pipeline to convert Exact Mapping Of Transcriptome Ends (EMOTE) data to the lists of quantified genomic positions correlated to related genomic information. J. Appl. Bioinform. Comput. Biol. 4. http://dx.doi.org/10.4172/2329-9533.1000118 (2015).
Acknowledgements
This work was supported by grant no. SNF#31003A_143660 to P.H.V. and a grant from the Société Académique de Genève to C.L.K. The authors acknowledge SNF/NCCR Chemical Biology for support of the chemical screening experiments and S. Kicka, V. Trofimov, D. Moreau and H. Riezman for advice and assistance with high-throughput screening. The authors thank P. Linder for discussions and funding contributions (SNF#31003A_149228/1) for the nEMOTE experiments, G. Panis for assistance with flow cytometry data acquisition and interpretation, C. Menck for the PimuA-lac290 and pNPTSΔlexA plasmids, L. Théraulaz for technical assistance, H. Yasrebi for nEMOTE data tabularization and J. Prados for assistance with R programming.
Author information
Authors and Affiliations
Contributions
C.L.K. and P.H.V. conceived and designed the study and wrote the manuscript. D.M. performed experiments (reported in Figs 1, 2 and Supplementary Figs 1–4) under the guidance of C.L.K. and P.H.V. P.R. designed, performed and analysed the nEMOTE experiments and wrote the corresponding sections of the manuscript. J.M. designed, performed and analysed the Tn-SEQ experiment. A.F. and J.B.C. contributed data analysis tools and analysed ChIP-SEQ and codon usage (A.F.) and high-throughput screening (J.B.C.) data. C.L.K., G.T., J.B.C. and M.C. designed the high-throughput screening experiments and C.L.K. performed these and all other experiments. M.C. and J.B.C. co-wrote the high-throughput screening section of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1–8, Note 1, Methods, Tables 1–4 and References. (PDF 1059 kb)
Supplementary Data 1
Analysis of LexA binding on the NA1000 chromosome by ChIPSEQ. (XLSX 67 kb)
Supplementary Data 2
Analysis of HigA binding on the NA1000 chromosome by ChIPSEQ. (XLSX 132 kb)
Supplementary Data 3
Validated HigB cleavage sites (5ʹ OH RNA ends) detected by nEMOTE. (XLS 154 kb)
Supplementary Data 4
Relative synonymous codon usage (RSCU) of the UCG-Ser codon per ORF in the NA1000 genome. (XLSX 220 kb)
Rights and permissions
About this article
Cite this article
Kirkpatrick, C., Martins, D., Redder, P. et al. Growth control switch by a DNA-damage-inducible toxin–antitoxin system in Caulobacter crescentus. Nat Microbiol 1, 16008 (2016). https://doi.org/10.1038/nmicrobiol.2016.8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.8