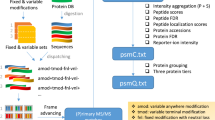
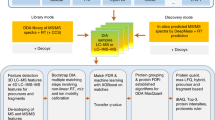
Abstract
Spectral searching has drawn increasing interest as an alternative to sequence-database searching in proteomics. We developed and validated an open-source software toolkit, SpectraST, to enable proteomics researchers to build spectral libraries and to integrate this promising approach in their data-analysis pipeline. It allows individual researchers to condense raw data into spectral libraries, summarizing information about observed proteomes into a concise and retrievable format for future data analyses.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Steen, H. & Mann, M. Nat. Rev. Mol. Cell Biol. 5, 699–711 (2004).
Eng, J.K., McCormack, A.L. & Yates, J.R., III. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994).
Patterson, S.D. Nat. Biotechnol. 21, 221–222 (2003).
Domon, B. & Aebersold, R. Mol. Cell. Proteomics 5, 1921–1926 (2006).
Yates, J.R., III, Morgan, S.F., Gatlin, C.L., Griffin, P.R. & Eng, J.K. Anal. Chem. 70, 3557–3565 (1998).
Domokos, L., Hennberg, D. & Weimann, B. Anal. Chim. Acta 165, 61–74 (1984).
Ausloos, P. et al. J. Am. Soc. Mass Spectrom. 10, 287–299 (1998).
Lam, H. et al. Proteomics 7, 655–667 (2007).
Craig, R., Cortens, J.C., Fenyo, D. & Beavis, R.C. J. Proteome Res. 5, 1843–1849 (2006).
Frewen, B.E., Merrihew, G.E., Wu, C.C., Noble, W.S. & MacCoss, M.J. Anal. Chem. 78, 5678–5684 (2006).
Keller, A., Eng, J., Zhang, N., Li, X.J. & Aebersold, R. Mol. Syst. Biol. 1, 17 (2005).
Pedrioli, P.G. et al. Nat. Biotechnol. 22, 1459–1466 (2004).
Deutsch, E.W. et al. Proteomics 5, 3497–3500 (2005).
Tabb, D.L., Thompson, M.R. & Khalsa-Moyers, G. J. Am. Soc. Mass Spectrom. 16, 1250–1261 (2005).
Flikka, K. et al. Proteomics 7, 3245–3258 (2007).
Acknowledgements
We thank all the data contributors to PeptideAtlas who have made this study possible. This study was supported in part by the US National Heart, Lung and Blood Institute, National Institutes of Health (N01-HV-28179).
Author information
Authors and Affiliations
Contributions
H.L. developed the SpectraST software, performed all library building and validation studies and wrote the manuscript. E.W.D. built the Human Plasma PeptideAtlas from which the data were drawn for this study. J.S.E., J.K.E. and S.E.S. contributed ideas to algorithm development and library format design. R.A. conceived and supervised the project. All authors discussed the results and commented on the manuscript.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Tables 1–4, Supplementary Methods (PDF 286 kb)
Rights and permissions
About this article
Cite this article
Lam, H., Deutsch, E., Eddes, J. et al. Building consensus spectral libraries for peptide identification in proteomics. Nat Methods 5, 873–875 (2008). https://doi.org/10.1038/nmeth.1254
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1254
This article is cited by
-
metaSpectraST: an unsupervised and database-independent analysis workflow for metaproteomic MS/MS data using spectrum clustering
Microbiome (2023)
-
Improving quantitation accuracy in isobaric-labeling mass spectrometry experiments with spectral library searching and feature-based peptide-spectrum match filter
Scientific Reports (2023)
-
A comprehensive spectral assay library to quantify the Halobacterium salinarum NRC-1 proteome by DIA/SWATH-MS
Scientific Data (2023)
-
Accurate de novo peptide sequencing using fully convolutional neural networks
Nature Communications (2023)
-
Inherited chitinases enable sustained growth and rapid dispersal of bacteria from chitin particles
Nature Microbiology (2023)