Abstract
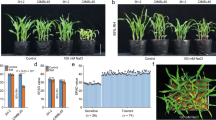
Many important agronomic traits in crop plants, including stress tolerance, are complex traits controlled by quantitative trait loci (QTLs). Isolation of these QTLs holds great promise to improve world agriculture but is a challenging task. We previously mapped a rice QTL, SKC1, that maintained K+ homeostasis in the salt-tolerant variety under salt stress1, consistent with the earlier finding that K+ homeostasis is important in salt tolerance2,3. To understand the molecular basis of this QTL, we isolated the SKC1 gene by map-based cloning and found that it encoded a member of HKT-type transporters. SKC1 is preferentially expressed in the parenchyma cells surrounding the xylem vessels. Voltage-clamp analysis showed that SKC1 protein functions as a Na+-selective transporter. Physiological analysis suggested that SKC1 is involved in regulating K+/Na+ homeostasis under salt stress, providing a potential tool for improving salt tolerance in crops.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Lin, H.X. et al. QTLs for Na+ and K+ uptake of shoot and root controlling rice salt tolerance. Theor. Appl. Genet. 108, 253–260 (2004).
Maathuis, F.J.M. & Amtmann, A. K+ nutrition and Na+ toxicity: the basis of cellular K+/Na+ ratios. Ann. Bot. (Lond.) 84, 123–133 (1999).
Qi, Z. & Spalding, E.P. Protection of plasma membrane K+ transport by the salt overly sensitive1 Na+/H+ antiporter during salinity stress. Plant Physiol. 136, 2548–2555 (2004).
Zhu, J.-K. Plant salt tolerance. Trends Plant Sci. 6, 66–71 (2001).
Akbar, M., Gunawardena, I.E. & Ponnamperuma, F.N. Breeding for soil stresses. in Progress in Rainfed Lowland Rice. (International Rice Research Institute, Manila, 1986).
Moons, A., Bauw, C., Prinsen, E., Montagu, M.V. & Straeten, D.V.D. Molecular and physiological responses to abscisic acid and salts in roots of salt-sensitive and salt-tolerant indica rice varieties. Plant Physiol. 107, 177–186 (1995).
Sasaki, T. et al. The genome sequence and structure of rice chromosome 1. Nature 420, 312–316 (2002).
Schachtman, D. & Liu, W. Molecular pieces to the puzzle of the interaction between potassium and sodium uptake in plants. Trends Plant Sci. 4, 281–287 (1999).
Schachtman, D.P. & Schroeder, J.I. Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature 370, 655–658 (1994).
Deckert, G. et al. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature 392, 353–358 (1998).
Uozumi, N. et al. The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis Oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol. 122, 1249–1259 (2000).
Garciadeblas, B., Senn, M.E., Banuelos, M.A. & Rodriguez-Navarro, A. Sodium transport and HKT transporters: the rice model. Plant J. 34, 788–801 (2003).
Kato, Y. et al. Evidence in support of a four transmembranepore-transmembrane topology model for the Arabidopsis thaliana Na+/K+ translocating AtHKT1 protein, a member of the superfamily of K+ transporters. Proc. Natl. Acad. Sci. USA 98, 6488–6493 (2001).
Wang, T.B., Gassmann, W., Rubio, F., Schroeder, J.I. & Glass, A.D.M. Rapid up-regulation of HKT1, a high-affinity potassium transporter gene, in roots of barley and wheat following withdrawal of potassium. Plant Physiol. 118, 651–659 (1998).
Berthomieu, P. et al. Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J. 22, 2004–2014 (2003).
Rubio, F., Gassmann, W. & Schroeder, J.I. Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270, 1660–1663 (1995).
Gassman, W., Rubio, F. & Schroeder, J.I. Alkali cation selectivity of the wheat root high-affinity potassium transporter HKT1. Plant J. 10, 869–882 (1996).
Horie, T. et al. Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. Plant J. 27, 129–138 (2001).
Liu, K. & Luan, S. Internal aluminum block of plant inward K+ channels. Plant Cell 13, 1453–1465 (2001).
Subramanian, V.S., Marchant, J.S., Parker, I. & Said, H.M. Intracellular trafficking/membrane targeting of human reduced folate carrier expressed in Xenopus oocytes. Am. J. Physiol. Gastrointest. Liver Physiol. 281, G1477–G1486 (2001).
Rus, A. et al. AtHKT1 facilitates Na+ homeostasis and K+ nutrition in planta. Plant Physiol. 136, 2500–2511 (2004).
Shi, H., Ishitani, M., Kim, C. & Zhu, J.-K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 97, 6896–6901 (2000).
Apse, M.P., Aharon, G.S., Snedden, W.A. & Blumwald, E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285, 1256–1258 (1999).
Epstein, E. The essential role of calcium in selective cation transport by plant cells. Plant Physiol. 36, 437–444 (1961).
Lander, E.S. & Botstein, D. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121, 185–199 (1989).
Mao, J., Zhang, Y.C., Sang, Y., Li, Q.H. & Yang, H.Q. A role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc. Natl. Acad. Sci. USA 102, 12270–12275 (2005).
Hiei, Y., Ohta, S., Komari, T. & Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282 (1994).
Lagarde, D. et al. Tissue-specific expression of Arabidopsis AKT1 gene is consistent with a role in K+ nutrition. Plant J. 9, 195–203 (1996).
Shi, H., Quintero, F.J., Pardo, J.M. & Zhu, J.-K. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14, 465–477 (2002).
Liman, E.R., Hess, P., Weaver, F. & Koren, G. Voltage-sensing residues in the S4 region of a mammalian potassium channel. Nature 353, 752–756 (1991).
Acknowledgements
We thank H.-Q. Yang for providing pHB vector, J. Yang, X.-Y. Huang and J.-J. Zhang for technical assistance and E.Y. Isacoff for providing X. laevis oocytes. This work was supported by grants from the Ministry of Science and Technology of China, the National Natural Science Foundation of China and the Shanghai Science and Technology Development Fund to H.-X.L. and a grant from the US Department of Agriculture to S.L.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Shoots Na+ contents (means±SE, n=5) in T1 rice transformants under normal or salt stress condition (120 mM NaCl for 7 days). (PDF 32 kb)
Supplementary Fig. 2
Sequence alignments of SKC1. (PDF 56 kb)
Supplementary Fig. 3
K+ and Na+ contents in the phloem sap were not significantly different between NIL(SKC1) and Koshihikari under normal or stress condition (25 mM NaCl). (PDF 34 kb)
Supplementary Fig. 4
NIL(SKC1) seedlings are more tolerant to salt then Koshihikari under salt stress (125 mM NaCl for 32 days). (PDF 158 kb)
Supplementary Table 1
The molecular marker primers developed in this study and primers for SKC1 analysis. (PDF 10 kb)
Rights and permissions
About this article
Cite this article
Ren, ZH., Gao, JP., Li, LG. et al. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet 37, 1141–1146 (2005). https://doi.org/10.1038/ng1643
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1643
This article is cited by
-
Natural variation in ZmNAC087 contributes to total root length regulation in maize seedlings under salt stress
BMC Plant Biology (2023)
-
Jacalin-related lectin 45 (OsJRL45) isolated from ‘sea rice 86’ enhances rice salt tolerance at the seedling and reproductive stages
BMC Plant Biology (2023)
-
Receptor-Like Cytoplasmic Kinase STK Confers Salt Tolerance in Rice
Rice (2023)
-
Genome-wide association studies identify OsWRKY53 as a key regulator of salt tolerance in rice
Nature Communications (2023)
-
Marker-Assisted Introgression of the Salinity Tolerance Locus Saltol in Temperate Japonica Rice
Rice (2023)