Abstract
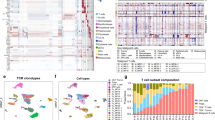
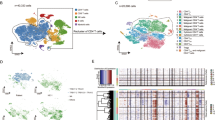
Sézary syndrome is a rare leukemic form of cutaneous T cell lymphoma characterized by generalized redness, scaling, itching and increased numbers of circulating atypical T lymphocytes. It is rarely curable, with poor prognosis. Here we present a multiplatform genomic analysis of 37 patients with Sézary syndrome that implicates dysregulation of cell cycle checkpoint and T cell signaling. Frequent somatic alterations were identified in TP53, CARD11, CCR4, PLCG1, CDKN2A, ARID1A, RPS6KA1 and ZEB1. Activating CCR4 and CARD11 mutations were detected in nearly one-third of patients. ZEB1, encoding a transcription repressor essential for T cell differentiation, was deleted in over one-half of patients. IL32 and IL2RG were overexpressed in nearly all cases. Our results demonstrate profound disruption of key signaling pathways in Sézary syndrome and suggest potential targets for new therapies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Campbell, J.J., Clark, R.A., Watanabe, R. & Kupper, T.S. Sézary syndrome and mycosis fungoides arise from distinct T-cell subsets: a biologic rationale for their distinct clinical behaviors. Blood 116, 767–771 (2010).
Girardi, M., Heald, P.W. & Wilson, L.D. The pathogenesis of mycosis fungoides. N. Engl. J. Med. 350, 1978–1988 (2004).
Kim, Y.H., Liu, H.L., Mraz-Gernhard, S., Varghese, A. & Hoppe, R.T. Long-term outcome of 525 patients with mycosis fungoides and Sézary syndrome: clinical prognostic factors and risk for disease progression. Arch. Dermatol. 139, 857–866 (2003).
Olsen, E. et al. Revisions to the staging and classification of mycosis fungoides and Sézary syndrome: a proposal of the International Society for Cutaneous Lymphomas (ISCL) and the cutaneous lymphoma task force of the European Organization of Research and Treatment of Cancer (EORTC). Blood 110, 1713–1722 (2007).
Herne, K.L., Talpur, R., Breuer-McHam, J., Champlin, R. & Duvic, M. Cytomegalovirus seropositivity is significantly associated with mycosis fungoides and Sézary syndrome. Blood 101, 2132–2136 (2003).
Talpur, R., Bassett, R. & Duvic, M. Prevalence and treatment of Staphylococcus aureus colonization in patients with mycosis fungoides and Sézary syndrome. Br. J. Dermatol. 159, 105–112 (2008).
Agar, N.S. et al. Survival outcomes and prognostic factors in mycosis fungoides/Sézary syndrome: validation of the revised International Society for Cutaneous Lymphomas/European Organisation for Research and Treatment of Cancer staging proposal. J. Clin. Oncol. 28, 4730–4739 (2010).
Scarisbrick, J.J. et al. Prognostic factors, prognostic indices and staging in mycosis fungoides and Sézary syndrome: where are we now? Br. J. Dermatol. 170, 1226–1236 (2014).
Talpur, R. et al. Long-term outcomes of 1,263 patients with mycosis fungoides and Sézary syndrome from 1982 to 2009. Clin. Cancer Res. 18, 5051–5060 (2012).
Vidulich, K.A., Talpur, R., Bassett, R.L. & Duvic, M. Overall survival in erythrodermic cutaneous T-cell lymphoma: an analysis of prognostic factors in a cohort of patients with erythrodermic cutaneous T-cell lymphoma. Int. J. Dermatol. 48, 243–252 (2009).
Kim, E.J. et al. Immunopathogenesis and therapy of cutaneous T cell lymphoma. J. Clin. Invest. 115, 798–812 (2005).
Ni, X., Zhang, C., Talpur, R. & Duvic, M. Resistance to activation-induced cell death and bystander cytotoxicity via the Fas/Fas ligand pathway are implicated in the pathogenesis of cutaneous T cell lymphomas. J. Invest. Dermatol. 124, 741–750 (2005).
Batista, D.A. et al. Multicolor fluorescence in situ hybridization (SKY) in mycosis fungoides and Sézary syndrome: search for recurrent chromosome abnormalities. Genes Chromosom. Cancer 45, 383–391 (2006).
Caprini, E. et al. Identification of key regions and genes important in the pathogenesis of Sézary syndrome by combining genomic and expression microarrays. Cancer Res. 69, 8438–8446 (2009).
Mao, X., Chaplin, T. & Young, B.D. Integrated genomic analysis of Sézary syndrome. Genet. Res. Int. 2011, 980150 (2011).
Mao, X. et al. Molecular cytogenetic characterization of Sézary syndrome. Genes Chromosom. Cancer 36, 250–260 (2003).
Vermeer, M.H. et al. Novel and highly recurrent chromosomal alterations in Sézary syndrome. Cancer Res. 68, 2689–2698 (2008).
Braun, F.C.M. et al. Tumor suppressor TNFAIP3 (A20) is frequently deleted in Sézary syndrome. Leukemia 25, 1494–1501 (2011).
Laharanne, E. et al. CDKN2A-CDKN2B deletion defines an aggressive subset of cutaneous T-cell lymphoma. Mod. Pathol. 23, 547–558 (2010).
Lamprecht, B. et al. The tumour suppressor p53 is frequently nonfunctional in Sézary syndrome. Br. J. Dermatol. 167, 240–246 (2012).
Vaqué, J.P. et al. PLCG1 mutations in cutaneous T-cell lymphomas. Blood 123, 2034–2043 (2014).
Duvic, M. & Foss, F.M. Mycosis fungoides: pathophysiology and emerging therapies. Semin. Oncol. 34, S21–S28 (2007).
Horwitz, S.M. Novel therapies for cutaneous T-cell lymphomas. Clin. Lymphoma Myeloma 8 (suppl. 5), S187–S192 (2008).
Venkatarajan, S. & Duvic, M. Sézary syndrome: an overview of current and future treatment options. Expert Opin. Orphan Drugs 2, 3 (2014).
Wang, L. et al. Novel somatic and germline mutations in intracranial germ cell tumours. Nature 511, 241–245 (2014).
Lawrence, M.S. et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014).
Wheeler, D.A. & Wang, L. From human genome to cancer genome: the first decade. Genome Res. 23, 1054–1062 (2013).
Alexandrov, L.B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Clark, R.A. Resident memory T cells in human health and disease. Sci. Transl. Med. 7, 269rv1 (2015).
Wu, X.S., Lonsdorf, A.S. & Hwang, S.T. Cutaneous T-cell lymphoma: roles for chemokines and chemokine receptors. J. Invest. Dermatol. 129, 1115–1119 (2009).
Yoshie, O. & Matsushima, K. CCR4 and its ligands: from bench to bedside. Int. Immunol. 27, 11–20 (2015).
Ni, X. et al. Reduction of regulatory T cells by Mogamulizumab, a defucosylated anti–CC chemokine receptor 4 antibody, in patients with aggressive/refractory mycosis fungoides and Sézary syndrome. Clin. Cancer Res. 21, 274–285 (2015).
Duvic, M. et al. Phase 1/2 study of mogamulizumab, a defucosylated anti-CCR4 antibody, in previously treated patients with cutaneous T-cell lymphoma. Blood 125, 1883–1889 (2015).
Nakagawa, M. et al. Gain-of-function CCR4 mutations in adult T cell leukemia/lymphoma. J. Exp. Med. 211, 2497–2505 (2014).
Matsumoto, R. et al. Phosphorylation of CARMA1 plays a critical role in T cell receptor–mediated NF-κB activation. Immunity 23, 575–585 (2005).
Hara, H. et al. The MAGUK family protein CARD11 is essential for lymphocyte activation. Immunity 18, 763–775 (2003).
Wang, D. et al. A requirement for CARMA1 in TCR-induced NF-κB activation. Nat. Immunol. 3, 830–835 (2002).
Lohr, J.G. et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl. Acad. Sci. USA 109, 3879–3884 (2012).
Lenz, G. et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science 319, 1676–1679 (2008).
Dereure, O., Portales, P., Clot, J. & Guilhou, J.J. Decreased expression of Fas (APO-1/CD95) on peripheral blood CD4+ T lymphocytes in cutaneous T-cell lymphomas. Br. J. Dermatol. 143, 1205–1210 (2000).
Jones, C.L. et al. Downregulation of Fas gene expression in Sézary syndrome is associated with promoter hypermethylation. J. Invest. Dermatol. 130, 1116–1125 (2010).
Ungewickell, A. et al. Genomic analysis of mycosis fungoides and Sézary syndrome identifies recurrent alterations in TNFR2. Nat. Genet. 47, 1056–1060 (2015).
Choi, J. et al. Genomic landscape of cutaneous T cell lymphoma. Nat. Genet. 47, 1011–1019 (2015).
Postigo, A.A. & Dean, D.C. Independent repressor domains in ZEB regulate muscle and T-cell differentiation. Mol. Cell. Biol. 19, 7961–7971 (1999).
Higashi, Y. et al. Impairment of T cell development in δEF1 mutant mice. J. Exp. Med. 185, 1467–1479 (1997).
Nakahata, S., Yamazaki, S., Nakauchi, H. & Morishita, K. Downregulation of ZEB1 and overexpression of Smad7 contribute to resistance to TGF-β1–mediated growth suppression in adult T-cell leukemia/lymphoma. Oncogene 29, 4157–4169 (2010).
Hidaka, T. et al. Down-regulation of TCF8 is involved in the leukemogenesis of adult T-cell leukemia/lymphoma. Blood 112, 383–393 (2008).
Blais, A. & Dynlacht, B.D. E2F-associated chromatin modifiers and cell cycle control. Curr. Opin. Cell Biol. 19, 658–662 (2007).
Nagl, N.G. Jr., Wang, X., Patsialou, A., Van Scoy, M. & Moran, E. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. EMBO J. 26, 752–763 (2007).
Nam, H.J. et al. The ERK-RSK1 activation by growth factors at G2 phase delays cell cycle progression and reduces mitotic aberrations. Cell. Signal. 20, 1349–1358 (2008).
Freeman, G.J. et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J. Exp. Med. 192, 1027–1034 (2000).
Okazaki, T., Chikuma, S., Iwai, Y., Fagarasan, S. & Honjo, T. A rheostat for immune responses: the unique properties of PD-1 and their advantages for clinical application. Nat. Immunol. 14, 1212–1218 (2013).
Suga, H. et al. The role of IL-32 in cutaneous T-cell lymphoma. J. Invest. Dermatol. 134, 1428–1435 (2014).
Ohmatsu, H. et al. IL32 is progressively expressed in mycosis fungoides independent of helper T-cell 2 and helper T-cell 9 polarization. Cancer Immunol. Res. 2, 890–900 (2014).
Kim, K.H. et al. Interleukin-32 monoclonal antibodies for immunohistochemistry, Western blotting, and ELISA. J. Immunol. Methods 333, 38–50 (2008).
Huether, R. et al. The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes. Nat. Commun. 5, 3630 (2014).
Vega, F. et al. Clonal heterogeneity in mycosis fungoides and its relationship to clinical course. Blood 100, 3369–3373 (2002).
Jackow, C.M. et al. Association of erythrodermic cutaneous T-cell lymphoma, superantigen-positive Staphylococcus aureus, and oligoclonal T-cell receptor Vβ gene expansion. Blood 89, 32–40 (1997).
Tan, R.S., Butterworth, C.M., McLaughlin, H., Malka, S. & Samman, P.D. Mycosis fungoides—a disease of antigen persistence. Br. J. Dermatol. 91, 607–616 (1974).
Bradford, P.T., Devesa, S.S., Anderson, W.F. & Toro, J.R. Cutaneous lymphoma incidence patterns in the United States: a population-based study of 3884 cases. Blood 113, 5064–5073 (2009).
Takashima, A. Establishment of fibroblast cultures. Curr. Protoc. Cell Biol. Ch. 2, Unit 2.1 (2001).
Lawrence, M.S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
DeLuca, D.S. et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 28, 1530–1532 (2012).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Roberts, A., Pimentel, H., Trapnell, C. & Pachter, L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27, 2325–2329 (2011).
McPherson, A. et al. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput. Biol. 7, e1001138 (2011).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Acknowledgements
We thank J. Hu, M. Wang, Y. Han, H. Chao and M.G. Evans for their excellent technical support. We thank L. Sandra for help with sample intake and J. Jayaseelan for project management. We thank W. Hale, D. Kalra, S. Dugan-Perez and J. Watt for their kind help with data submission. Special thanks is given to D. Burgess and M. Chase from Roche NimbleGen for their great help with the design and fast delivery of the custom capture array. This work was supported by research funding from the National Human Genome Research Institute (NHGRI; grant 5U54HG003273) and the Cancer Prevention Research Institute of Texas (CPRIT; grant RP121018) to D.A.W., the Drs. Martin and Dorothy Spatz Charitable Foundation (grant 00005840), the Blanche Bender Professorship in Cancer Research and the MD Anderson Cancer Center Core Grant (grant CA16672) to M.D.
Author information
Authors and Affiliations
Contributions
L.W. conducted the major bioinformatics analyses of the multiplatform data, and wrote and revised the manuscript. X.N. contributed to performing the research, data interpretation and manuscript preparation. K.R.C. contributed to mutation signature analysis. B.Y.Y. and J.S. contributed to performing the research. X.Z. collected tumor specimens and performed DNA and RNA extraction, and immunoblot assays. L.X. and J.D. contributed to the RNA-seq and whole-exome sequencing pipeline. Q.M. contributed to the fusion gene validation experiments. T.L. helped with the serum and ELISA assays. D.M.M. and H.D. managed the production pipeline of exome, RNA-seq and SNP array analysis. L.A.D. contributed the somatic mutation significance analysis. R.A.G. contributed to revision of the manuscript. D.A.W., M.D. and L.W. conceived the study and supervised the research. D.A.W. and M.D. also contributed to the writing and revision of the manuscript. M.D. recruited, consented, staged, characterized and cared for the patients and supervised the skin biopsies.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12. (PDF 8255 kb)
Supplementary Tables 1–17
Supplementary Tables 1–17. (XLSX 3207 kb)
Rights and permissions
About this article
Cite this article
Wang, L., Ni, X., Covington, K. et al. Genomic profiling of Sézary syndrome identifies alterations of key T cell signaling and differentiation genes. Nat Genet 47, 1426–1434 (2015). https://doi.org/10.1038/ng.3444
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3444
This article is cited by
-
Naturally occurring T cell mutations enhance engineered T cell therapies
Nature (2024)
-
MHC-I upregulation safeguards neoplastic T cells in the skin against NK cell-mediated eradication in mycosis fungoides
Nature Communications (2024)
-
SWI/SNF complexes in hematological malignancies: biological implications and therapeutic opportunities
Molecular Cancer (2023)
-
Primary pulmonary T-cell lymphoproliferative disorders with a limited-stage, low proliferative index, and unusual clinical behavior: two cases of a rare occurrence
Virchows Archiv (2023)
-
Integrated clinical genomic analysis reveals xenobiotic metabolic genes are downregulated in meningiomas of current smokers
Journal of Neuro-Oncology (2023)