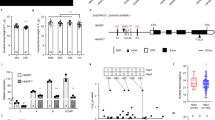
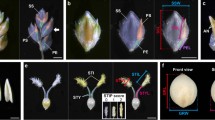
Abstract
Domestication of cereal crops, such as maize, wheat and rice, had a profound influence on agriculture and the establishment of human civilizations. One major improvement was an increase in seed number per inflorescence, which enhanced yield and simplified harvesting and storage1,2. The ancestor of maize, teosinte, makes 2 rows of kernels, and modern varieties make ∼8–20 rows3. Kernel rows are initiated by the inflorescence shoot meristem, and shoot meristem size is controlled by a feedback loop involving the CLAVATA signaling proteins and the WUSCHEL transcription factor4,5. We present a hypothesis that variation in inflorescence meristem size affects kernel row number (KRN), with the potential to increase yield. We also show that variation in the CLAVATA receptor–like protein FASCIATED EAR2 leads to increased inflorescence meristem size and KRN. These findings indicate that modulation of fundamental stem cell proliferation control pathways has the potential to enhance crop yields.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Bommert, P., Satoh-Nagasawa, N., Jackson, D. & Hirano, H.Y. Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol. 46, 69–78 (2005).
Evans, L.T. Crop Evolution, Adaptation and Yield (Cambridge University Press, Cambridge, 1993).
Doebley, J. The genetics of maize evolution. Annu. Rev. Genet. 38, 37–59 (2004).
Brand, U., Fletcher, J.C., Hobe, M., Meyerowitz, E.M. & Simon, R. Dependence of stem cell fate in Arabidopsis an a feedback loop regulated by CLV3 activity. Science 289, 617–619 (2000).
Schoof, H. et al. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644 (2000).
Gregory, P.J. & George, T.S. Feeding nine billion: the challenge to sustainable crop production. J. Exp. Bot. 62, 5233–5239 (2011).
Frary, A. et al. fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289, 85–88 (2000).
Jiao, Y. et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544 (2010).
Miura, K. et al. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42, 545–549 (2010).
Ashikari, M. et al. Cytokinin oxidase regulates rice grain production. Science 309, 741–745 (2005).
Huang, X. et al. Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 41, 494–497 (2009).
Li, Y. et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43, 1266–1269 (2011).
Barton, M.K. Twenty years on: the inner workings of the shoot apical meristem, a developmental dynamo. Dev. Biol. 341, 95–113 (2010).
Ha, C.M., Jun, J.H. & Fletcher, J.C. Shoot apical meristem form and function. Curr. Top. Dev. Biol. 91, 103–140 (2010).
Komatsuda, T. et al. Six-rowed barley originated from a mutation in a homeodomain–leucine zipper I–class homeobox gene. Proc. Natl. Acad. Sci. USA 104, 1424–1429 (2007).
Ramsay, L. et al. INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nat. Genet. 43, 169–172 (2011).
Lu, M. et al. Mapping of quantitative trait loci for kernel row number in maize across seven environments. Mol. Breed. 28, 143–152 (2011).
Upadyayula, N., da Silva, H.S., Bohn, M.O. & Rocheford, T.R. Genetic and QTL analysis of maize tassel and ear inflorescence architecture. Theor. Appl. Genet. 112, 592–606 (2006).
Veldboom, L.R., Lee, M. & Woodman, W.L. Molecular marker–facilitated studies in an elite maize population. 1. Linkage analysis and determination of QTL for morphological traits. Theor. Appl. Genet. 88, 7–16 (1994).
Lee, M. et al. Expanding the genetic map of maize with the intermated B73 × Mo17 (IBM) population. Plant Mol. Biol. 48, 453–461 (2002).
Wang, S., Basten, C.J. & Zeng, Z.B. Windows QTL Cartographer 2.5 (Department of Statistics, North Carolina State University, Raleigh, North Carolina, 2011).
Taguchi-Shiobara, F., Yuan, Z., Hake, S. & Jackson, D. The fasciated ear2 gene encodes a leucine-rich repeat receptor–like protein that regulates shoot meristem proliferation in maize. Genes Dev. 15, 2755–2766 (2001).
Jeong, S., Trotochaud, A.E. & Clark, S.E. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor–like kinase. Plant Cell 11, 1925–1934 (1999).
Fiers, M. et al. The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 17, 2542–2553 (2005).
Guo, Y., Han, L., Hymes, M., Denver, R. & Clark, S.E. CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. Plant J. 63, 889–900 (2010).
McMullen, M.D. et al. Genetic properties of the maize nested association mapping population. Science 325, 737–740 (2009).
Till, B.J. et al. Discovery of induced point mutations in maize genes by TILLING. BMC Plant Biol. 4, 12 (2004).
Weil, C.F. TILLING in grass species. Plant Physiol. 149, 158–164 (2009).
Ng, P.C. & Henikoff, S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814 (2003).
DeYoung, B.J. et al. The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase–like proteins are required for meristem function in Arabidopsis. Plant J. 45, 1–16 (2006).
Deyoung, B.J. & Clark, S.E. BAM receptors regulate stem cell specification and organ development through complex interactions with CLAVATA signaling. Genetics 180, 895–904 (2008).
Müller, R., Bleckmann, A. & Simon, R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell–limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20, 934–946 (2008).
Kinoshita, A. et al. RPK2 is an essential receptor–like kinase that transmits the CLV3 signal in Arabidopsis. Development 137, 3911–3920 (2010).
Betsuyaku, S. et al. Mitogen-activated protein kinase regulated by the CLAVATA receptors contributes to shoot apical meristem homeostasis. Plant Cell Physiol. 52, 14–29 (2011).
Bommert, P. et al. thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor–like kinase. Development 132, 1235–1245 (2005).
Suzaki, T. et al. The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development 131, 5649–5657 (2004).
Kinoshita, T. et al. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433, 167–171 (2005).
Ogawa, M., Shinohara, H., Sakagami, Y. & Matsubayashi, Y. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319, 294 (2008).
She, J. et al. Structural insight into brassinosteroid perception by BRI1. Nature 474, 472–476 (2011).
Jaillais, Y., Belkhadir, Y., Balsemão-Pires, E., Dangl, J.L. & Chory, J. Extracellular leucine-rich repeats as a platform for receptor/coreceptor complex formation. Proc. Natl. Acad. Sci. USA 108, 8503–8507 (2011).
Nimchuk, Z.L., Tarr, P.T. & Meyerowitz, E.M. An evolutionarily conserved pseudokinase mediates stem cell production in plants. Plant Cell 23, 851–854 (2011).
Müller, R., Borghi, L., Kwiatkowska, D., Laufs, P. & Simon, R. Dynamic and compensatory responses of Arabidopsis shoot and floral meristems to CLV3 signaling. Plant Cell 18, 1188–1198 (2006).
Nimchuk, Z.L., Tarr, P.T., Ohno, C., Qu, X. & Meyerowitz, E.M. Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Curr. Biol. 21, 345–352 (2011).
Brown, P.J. et al. Distinct genetic architectures for male and female inflorescence traits of maize. PLoS Genet. 7, e1002383 (2011).
Shepard, K.A. & Purugganan, M.D. Molecular population genetics of the Arabidopsis CLAVATA2 region. The genomic scale of variation and selection in a selfing species. Genetics 163, 1083–1095 (2003).
Muños, S. et al. Increase in tomato locule number is controlled by two single-nucleotide polymorphisms located near WUSCHEL. Plant Physiol. 156, 2244–2254 (2011).
Bomblies, K. & Doebley, J.F. Pleiotropic effects of the duplicate maize FLORICAULA/LEAFY genes zfl1 and zfl2 on traits under selection during maize domestication. Genetics 172, 519–531 (2006).
Searle, S.R. Linear Models (John Wiley & Sons, New York, 1971).
Hallauer, A. & Miranda, J. Quantitative Genetics in Maize Breeding (Iowa State University Press, Ames, Iowa, 1985).
Knapp, S.J., Stroup, W.W. & Ross, W.M. Exact confidence intervals for heritability on a progeny mean basis. Crop Sci. 25, 192–194 (1985).
Utz, H.F. PLABSTAT. Ein Computerprogramm zur Statistischen Analyse von Pflanzenzüchterischen Experimenten (Universität Hohenheim, Stuttgart, Germany, 1998).
Doerge, R.W. & Churchill, G.A. Permutation tests for multiple loci affecting a quantitative character. Genetics 142, 285–294 (1996).
Acknowledgements
We wish to thank C. Weil and the Maize TILLING facility for the isolation of the fea2 TILLING alleles, F. Taguchi Shiobara for SEM images, U. Au, L.A. Haller and K. Lau for KRN counting, P. Yin and K. Chen for assistance with genotyping, M. Bohn and S. Goldshmidt for assistance with statistical analysis, members of the Jackson laboratory for comments on the manuscript, T. Rocheford for growing IBM recombinant inbred lines, M. Komatsu, L. Heetland, K. Simcox and H. Sakai (DuPont- Pioneer) for grow-outs of TILLING populations and yield trait measurements, P. Brown and E. Buckler for discussions and A. Eveland for informatics. Funding from the US Department of Agriculture (USDA; grant NRICGP 2003-35304-13277), the National Science Foundation (NSF) Plant Genome Program (grant DBI-0604923) and the German Science Society (DFG) for a postdoctoral fellowship (grant Bo 3012/1-1) to P.B. is gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
P.B. performed all experiments, except where noted below, analyzed the data and helped with writing the manuscript. N.S.N. generated the data on the variation in maize inflorescence meristem size in different inbred lines. D.J. assisted with data analysis and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–3 and 5 (PDF 289 kb)
Supplementary Table 4
Genes and gene density in the FEA2 QTL interval. See separate file: FEA2_interval_gene annotations.xls (XLS 60 kb)
Rights and permissions
About this article
Cite this article
Bommert, P., Nagasawa, N. & Jackson, D. Quantitative variation in maize kernel row number is controlled by the FASCIATED EAR2 locus. Nat Genet 45, 334–337 (2013). https://doi.org/10.1038/ng.2534
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2534
This article is cited by
-
Characterization of sub-tropical maize (Zea Mays L.) inbred lines for the variation in kernel row numbers (KRNs)
Cereal Research Communications (2024)
-
Profiling the selected hotspots for ear traits in two maize–teosinte populations
Theoretical and Applied Genetics (2024)
-
Identification and validation of plant height, spike length and spike compactness loci in common wheat (Triticum aestivum L.)
BMC Plant Biology (2022)
-
GIF1 controls ear inflorescence architecture and floral development by regulating key genes in hormone biosynthesis and meristem determinacy in maize
BMC Plant Biology (2022)
-
Systematic trait dissection in oilseed rape provides a comprehensive view, further insight, and exact roadmap for yield determination
Biotechnology for Biofuels and Bioproducts (2022)