Abstract
Grains and glasses, widely different materials, arrest their motions upon decreasing temperature and external load, respectively, in common ways, leading to a universal jamming phase diagram conjecture. However, unified theories are lacking, mainly because of the disparate nature of the particle interactions. Here we demonstrate that folded proteins exhibit signatures common to both glassiness and jamming by using temperature- and force-unfolding molecular dynamics simulations. Upon folding, proteins develop a peak in the interatomic force distributions that falls on a universal curve with experimentally measured forces on jammed grains and droplets. Dynamical signatures are found as a dramatic slowdown of stress relaxation upon folding. Together with granular similarities, folding is tied not just to the jamming transition, but a more nuanced picture of anisotropy, preparation protocol and internal interactions emerges. Results have implications for designing stable polymers and can open avenues to link protein folding to jamming theory.
Similar content being viewed by others
Introduction
Granular matter (such as sand, coal or powder) is rigid when jammed, but flows when external loads are applied. Experiments show that jammed grains resist flow by forces that are propagated through the material across percolating networks of immovable particles, complex sets of force chains that give rise to rich phenomenology1,2. Different in nature from grains, glasses, obtained from supercooled liquids that avoid crystalization3, are also rigid when cold, but flow when their temperature is increased. Grains and glasses seem to ‘unjam’ alike; the physical change they have in common is a variation in their mechanical response to external perturbations from a stable, disordered configuration to a flowing, loose state. Common behaviour has been suggested to exist in the form of a ‘universal jamming phase diagram’4 mapping the arrest of dynamics in all such systems (see Fig. 1a). This commonality is despite the fundamentally distinct nature of the interactions involved: for glasses, which are atomic systems, the inter-particle energies are comparable to kBT and unjamming occurs due to thermal effects, whereas granular matter belongs to a completely different class. The characteristic interactions between grains are so large that temperature is irrelevant and energy dissipates almost instantaneously through friction (making difficult—except for simple models5—a traditional statistical mechanical approach). For the athermal grains, unjamming occurs because of the external field applied. In the wake of the conjecture of the universality of the jamming diagram, phenomenology common to jammed grains6 and supercooled systems has been sought and reported increasingly frequently5,7,8,9,10,11,12,13, in an effort to unify glass and jamming transitions. However, a common theory for the origins of the assumed unification is lacking, chiefly because the interactions responsible for the dynamical arrest are so different. More importantly, detailed new experiments on grains have revealed that there are differences even within the granular realm itself; under a variety of conditions, a more complex picture of jamming than that sketched in the original jamming diagram is emerging. This includes the intricacies of the preparation protocol, which can induce granular jamming into several distinct states14, a more subtle role of shear than originally envisaged15,16,17, and the importance of frictional interactions18. A more complex jamming scenario, rather than a single, overarching universality, is also revealed when considering criticality arguments; upon approaching a critical value Φc of the jamming order parameter Φ (for example, the packing density), zero temperature granular matter develops correlations that diverge like |Φ−Φc|−γ (refs 19,20). But unlike universal power law divergence around thermodynamical critical points, in granular matter, depending on the shearing conditions, different groups (working on different systems and/or conditions) find different critical exponents γ (refs 21,22,23).
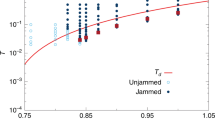
(a) Schematic phase diagram for ‘unified jamming’ adapted from ref. 4 to proteins. Below certain values of external force (Fext), temperature (T) and denaturant concentration (Φ, or other environmental parameter), proteins fold and undergo changes in mechanical properties similar to jamming of grains. (b,c) Caricatural analogy between shearing jammed grains and unfolding proteins: (b) jammed grains: black beads carry stress balancing paths, yellow depicts less stress bearing beads, and blue beads are least stressed, loose grains (cf. Cates et al.2). Force chains marked with red lines; black arrows indicate direction of shear. (c) Proteins pulled by external forces (in analogy with panel b), with backbone as the force transmission path (colour coding of atoms-beads correspondence as in b); the dotted lines depict non-bonding interactions. (d,e) Radius of gyration of proteins versus T and Fext; a sudden increase of Rg defines the transition values of T and Fext where proteins loose majority of native contacts and ‘unjam’.
Important clues have emerged from experiments on grains and colloids, and from simulations of glasses. For grains approaching jamming, a signature of the transition develops in the form of a measurable peak in the low-force region of the distribution of contact force components24,25,26,16, a peak non-existent in the loose, unjammed state. Simulations of grains as frictionless particles confirmed the onset of such a peak on jamming27. In the realm of glasses, atomistic simulations of glass-forming liquids also have shown that a peak appears in the inter-particle normal force near7,9 or, in one case, much below8 the glass transition temperature Tg and have suggested that the peak is related to the fragility3 of the glass former. On the other hand, in the realm of grains, ‘fragility’ refers to states that are robust under continued shear, but that unjam if the shear changes direction. This fragile response was linked2 to the marginal stability of the percolating network of force chains1 within the material (see Fig. 1b).
Concomitant with results on several granular systems for which a peak in the force distribution was deemed symptomatic of jamming, important subtleties have been observed. The force distribution can have a more complex connection to the state of the system. For example, the shape of the peak for jammed systems is sensitive to whether the system is sheared (that is, if external load is anisotropic) or not15,16,28. At the same time, the long f-tails are functionally sensitive to external stress conditions (see below).
Although a jamming peak appears in both grains and glasses, temperature-controlled and load-controlled jamming are usually observed in different systems, because it is difficult to apply forces to temperature-dependent glasses and it is difficult to define a temperature for grains under load. Therefore, with a few notable exceptions, such as a study of attractive colloids under external stress6 and the rheology of vibrated disks29,30 (which however—even when the vibration balances dissipation—are in non-equilibrium steady states at best, and do not truly reach thermal equilibrium, but thermodynamic analogies of which are nonetheless intriguing31), systems for which jamming occurs collectively across the T−Fext plane (Fig. 1a) are scarce, because when acting naturally Fext and T are applied in different systems (throughout the paper, we denote by Fext the external force applied on the system to distinguish it from internal forces F on each particle).
Here we suggest that proteins are systems that naturally populate the T−Fext plane and aim to describe their folded states using the language and descriptors of jamming. In the process, we reveal parallels, but also differences, between the protein-folding transition and the jamming transition. The study is of relevance, because proteins are systems whose dynamics and thermodynamics are naturally modulated by both the temperature and external force. First, increasing temperature above a certain folding temperature induces reversible protein unfolding32; this temperature-controlled change is akin to a glass-to-liquid transition. As such, rich theoretical advancement has been made for protein folding using an energy landscape description inspired by glasses33,34. Second, many proteins, such as those translocated through membranes or those degraded by ATP-dependent proteases, undergo regulated unfolding in vivo under the influence of externally applied forces35. Proteins are also unfolded in vitro by single-molecule pulling with atomic force microscopy, optical or magnetical tweezers to study the heterogeneity of their unfolding pathways36; under such external loads, signatures of glassy dynamics are again manifested37.
Results
Temperature and force unfolding of proteins
We chose proteins calmodulin, titin (the I27 module) and ubiquitin from the three major types of secondary structure combinations: all α-helical, all β-sheet and mixed, respectively. The three proteins were simulated using molecular dynamics simulations38 in their native, folded states, as well as at increasing temperatures, T, or under increasing external forces, Fext, applied at the two ends (all at T=300 K) to induce protein unfolding. We used the variation of their radius of gyration Rg as a gauge of the degree of protein unfolding (Fig. 1d). Samples from the various temperature- and force-dependent molecular dynamics trajectories were used to generate structural ensembles of folded or unfolded structures (depending on the value of T or Fext). With these ensembles, we then set to explore how variations in the interatomic force distributions correlate with the folding–unfolding transition. Moreover, proteins are macromolecules with a well-defined connectivity along the bonded backbone, which can act, on the application of external forces, as stress transmission paths that are heuristically comparable to force chains in granular materials (compare Fig. 1b with c). As in any given protein the same interactions are responsible for both the temperature- and force-induced unjamming, we set to directly compare which protein microscopic signatures may, at the onset of jamming, be common with granular-like and glass-like matter.
Given that in their thermal unfolding proteins contain vestiges of the glass-to-liquid (that is, unjamming) transition, it is tempting to regard protein unfolding by external pulling at their ends in similarity to unjamming grains by external shear force. However, complexities arise. Stress chains fail in sheared dry non-cohesive granular material because of compression in the chain direction and by a process akin to buckling. The physics of buckling under compression and that of failure by extension, the later being the process for the present study, are generally not the same. In the case of proteins, unjamming under extension may correspond to a different failure mode. The analogue of the pulling scenario for a granular system would require cohesive grains (cf. Fig. 1b), and such a system might have a different phenomenology worth testing in its own right, for example, in attractive colloidal suspensions6 (also, see below the discussion on force pulling). We note that graphs like the schematic universal phase diagram were produced as a result of using lattice and off-lattice models to understand the collective effect of external force and temperature, and temperature softening and related ideas were discussed (see refs 39,40).
Signatures in the distribution of forces
Using ingenious measurements, experiments on jammed grains41 and colloids42 have recorded entire distributions of single-particle forces, which reveal a peak upon jamming as discussed. To compare with these experimental data, we have computed from our simulations F=|−∂V/∂ri|,∀i, that is, the force magnitude on any single particle i due to all terms in the potential energy function V({ri}). We then normalized forces in all cases (simulation and experiment) by f=F/〈F〉, with 〈F〉 the average force over all force data for the corresponding system (proteins, grains, colloids). Despite the span of nine orders of magnitude between atomic and grain forces, an absolutely remarkable universal peak (Fig. 2) became apparent in all the distributions P(f) of scaled forces for grains, colloids and the three proteins.
To directly compare with structural signatures of jammed glasses7, whose atomic interactions have an important long-range component, we have computed the non-bonded contribution to the force distribution, Pnb(fnb), where the scaled forces were fnb=Fnb/〈Fnb〉, with  , being the magnitude of the force on any atom i from non-bonded pair interactions
, being the magnitude of the force on any atom i from non-bonded pair interactions  (electrostatic and van der Waals) with all the other atoms j.
(electrostatic and van der Waals) with all the other atoms j.
In Fig. 3, the distribution Pnb(fnb) also shows a peak when the proteins are folded, as already shown for P(f). As for P(f), the height of the peak in Pnb(fnb) decreases as T or Fext increase; this reflects a decrease in the degree of jamming. Unlike the universal peak in P(f), however, the size of the peak in Pnb(fnb) is protein-dependent, being more pronounced for more mechanically stable proteins. For the curves plotted in Fig. 3, the dependence Pnb(fnb) around the peak can be fitted to a function of the form
Peak disappearance upon temperature and force unfolding for the three proteins studied. Semi-log plot of force distribution Pnb(fnb) for non-bonding interactions against scaled force fnb. Variation of Pnb(fnb) under different Ts shown in a,c,e, and under different Fext in b,d,f for titin (a,b), ubiquitin (c,d) and calmodulin (e,f). For high Fext forces (that is, beyond unfolding, for example, green curve in Fig. 3b), note the additional ‘ripples’ appearing for small fnb in accord with data on triangular lattices under increasing anisotropy of shear (cf., for example, Fig. 5 in ref. 46 and references therein). Variation of exponent α (a measure of peak size) against T in panel g, and α against Fext in h. The magnitude of α (see text) correlates with the mechanical unfolding resilience and may be considered as a measure of the amount of stress stored in the non-bonding interactions. Titin, which has highest value of α, is one of the strongest proteins found in nature.

with f0 and A constants, and the fitting parameter α >1 giving a measure of peak size. The variation of α correlates with the perception of mechanical resilience to unfolding. Values of α close to unity (that is, no peak) are seen in the unjammed states, whereas the maximum value of α (largest peak) corresponds to the maximally jammed state (that is, at null Fext and room temperature) of the maximally stable protein. Figure 3g ranks titin highest in stability, stability that decreases as T increases; ubiquitin is next and calmodulin is least stable; the same order of mechanical resilience was found in the single molecule pulling experiments43,44,45. Figure 3h shows that this α-ordering holds for our force-dependent unfolding data, too; under Fext, α is a measure of the increase of jamming. Near the sudden break of the folded structure, α decreases for small increments of Fext. On further increases of Fext, unfolding (that is, the jamming to unjamming transition) ensues. Our results suggest that an analysis of Pnb(fnb) in the folded state of a wide range of proteins, with a tabulation of the respective α-values, may have predictive power in ranking mechanical or thermal stability.
Moreover, it is known that, depending on the protocol for minimization during, for example, simulated annealing, proteins can be trapped in metastable states around the folded state. This is also analogous to the dependence of the glass transition temperature on the cooling rate in glasses. We expect that the distributions Pnb(fnb) of trapped intermediates can feature peaks similar to the folded state, but with α-values correlating to stability. Jamming in frictionless granular systems also can feature a multiplicity of jammed states surrounding the frictionless point in the phase diagram14, accessing each requiring a different preparation protocol. An analogy to such protocol-dependent, distinct jammed states might be present in studies of metastable conformational states in proteins.
Although the peak fits the super-exponential form in equation 1, that is, with α >1) in the low-force region (around f≈1), the force distribution of the pair forces Pnb(fnb) at the high-force end shows a stretched exponential tail,  , with β <1 in the folded state. Analysis of individual-type interaction
, with β <1 in the folded state. Analysis of individual-type interaction  (that is, of each interaction between the same pair type of atoms i and j) revealed an exponential pair-force distribution tail (in accord with the simulations of Lennard–Jones systems) and differences in the peak shape and position (see Supplementary Fig. S1). The decay constant f0 of each of the tails for different atom pairs are however different; thus, their superposition leads to a stretched exponential. Upon unfolding, β→1 because a limited number of interaction types exists within the non-bonded cut-off and through the backbone in the unfolded state; this narrows considerably the distribution of characteristic decay forces extant in the folded state and stretched exponentiality hence subsides.
(that is, of each interaction between the same pair type of atoms i and j) revealed an exponential pair-force distribution tail (in accord with the simulations of Lennard–Jones systems) and differences in the peak shape and position (see Supplementary Fig. S1). The decay constant f0 of each of the tails for different atom pairs are however different; thus, their superposition leads to a stretched exponential. Upon unfolding, β→1 because a limited number of interaction types exists within the non-bonded cut-off and through the backbone in the unfolded state; this narrows considerably the distribution of characteristic decay forces extant in the folded state and stretched exponentiality hence subsides.
Comparison of pairwise unscaled force distributions P(Fij) in equilibrium with thermally and force-unfolded states (Fig. 4a), respectively, shows that the peak centres ∼5–15 pN in the folded state and that it disappears in both scenarios of unfolding. Note (cf. inset to Fig. 4a) that the protein peak is the region where contributions from the bonding and non-bonding components of P(Fij) are comparable (bonding interactions dominate P(Fij) in the high-force end, whereas the non-bonding interactions dominate in the low-force region). On the other hand, there is no peak disappearing in the bonding distribution upon either T- or Fext-unjamming. This indicates that the peak arises from the non-bonding interactions that are transmitted through the bonded structure (the skeleton) of the protein, that serves as a force chain for jamming. We also note, making again reference to the inset of Fig. 4a, that, over the low-force region, P(Fij) is scale-free for non-bonding interactions, decaying as a power law  . Interestingly, the value of γ is the same; it approximates 1.6 for all three proteins in the folded state studied herein and 1.5 in the unfolded state in all trajectories.
. Interestingly, the value of γ is the same; it approximates 1.6 for all three proteins in the folded state studied herein and 1.5 in the unfolded state in all trajectories.
(a) Pair-force distribution P(Fij) for ubiquitin for bonding, non-bonding and all-type interactions are compared between two states: equilibrium folded state at 300 K (jammed) and thermally unfolded state at T=700 K (T-unjammed). Probability density between 5 and 15 pN, (that is, the peak) disappears upon both force- and temperature-induced unfolding for non-bonding forces only. Inset of a shows log–log plot of P(Fij) for the full range of forces; non-bonding interactions prevail in the low-force region and bonding interactions prevail at the high-force end. (b) Comparison of P(Fij) for folded state at 300 K without external force (jammed) and force-unfolded state at Fext=1,000 pN (Fext-unjammed). (c) The distribution P(Fnb) of total non-bonded force on each atom is compared for: jammed (folded) state at 300 K, thermally unjammed at T=700 K and unjammed under external force Fext=1,000 pN. Inset of c shows corresponding force distribution of total force on each atom from all contributions (bonding and non-bonding) over the entire force range.
Showcasing ubiquitin as an example (titin and calmodulin show similar profiles and characteristic peak forces (see Supplementary Fig. S2), Fig. 4c shows that the force distribution of the sum of the forces on each atom from non-bonding interactions P(Fnb) also develops a peak for the folded state relative to the distributions recorded upon thermal or force-induced unfolding. The inset to Fig. 4c shows the distribution P(F) of the total forces on each atom from all contributions (bonding and non-bonding) over the entire spread of forces in the folded versus unfolded state. Comparison of Fig. 4c with its inset shows that, when the protein unfolds the non-bonding force distribution contracts, whereas the all-type contribution (that is, from bonding plus non-bonding forces) spreads; this is attributed to the fact that when the protein is unfolded, stress relaxation through non-bonding interactions reduces. In the inset (Fig. 4c), the all-type distribution P(F) for unjamming under a force so high that it pulls the protein taut (Fext=1,000 pN) coincides with the T=300 K jammed (folded) state. This is because in the distribution P(F) of the total forces F, the major contribution is from bonding interactions (contribution from non-bonding interactions are negligible). The force distribution of the bonding interaction varies significantly only with temperature variation; it shows negligible changes upon force unjamming at the same temperature.
So taken together, where do our results on protein force distributions stand in relationship to the universal jamming diagram? Is protein folding in a universality class with grains, glasses and other systems undergoing dynamical arrest? The answer is complex. The conjecture by O’Hern et al.7 has put forth that the peak in P(f) is symptomatic of jamming. Our results herein also show that a peak appears in folded proteins. However, even for grains, several subtleties have been revealed regarding the overall shape of P(f) (that is, peak and tail) and its relation to jamming. Evidence is accumulating from several granular studies that both the peak shape and the large f-tail of the distribution are strongly influenced by anisotropy, not just by (un)jamming. The effect of anisotropy on the peak was explored theoretically15,28 (see also review by Tighe et al.46). For small f, rippling in P(f) has been shown in frictionless triangular lattices under increasingly anisotropic external shear stress (compare, for example, Fig. 5 in ref. 46 with ripples for high Fext, for example, green curves in our Fig. 3b). Experimentally also, the peak was shown to be different in purely sheared versus uniformly compressed grains16. The same experiments also revealed changes in the tail of P(f), with theory46 showing an exponential character in the tails evolving as the shear stress/normal stress grows. This leaves the exact shape of P(f) still an open question for granular materials, and this richness in details is reflected by our results in the protein realm.
Time-dependence of stress relaxation
Another characteristic property of jammed granular matter is the slowing down of stress relaxation47,48, a dynamic signature resembling the viscous slowdown of glasses3. Given the commonality of the jamming peak in the force distributions across folded proteins, grains and glasses, we expected that whenever jamming in the conventional sense occurs in a protein (that is, when folding occurs) stress relaxation throughout the protein should also slow down, relative to the unfolded state.
To report on the mechanical stress at the atomic scale, we used the internal virial stress tensor  , where μ and ν label the Cartesian components x, y, z of the position and particle force vectors, respectively, and the summation is the overall atoms. We also computed the non-bonded contribution to the virial,
, where μ and ν label the Cartesian components x, y, z of the position and particle force vectors, respectively, and the summation is the overall atoms. We also computed the non-bonded contribution to the virial,  , with
, with  and
and  being the ν and μ components of the non-bonding pair force and distance, respectively, between two atoms i and j. We gauged stress relaxation by the decay of the normalized stress–stress time correlation function S(t)=(C(t)−C(∞))/(C(0)−C(∞)), where C(t)=〈σμν(t)σμν(0)〉. The correlations S(t) in all directions did slow down in the folded state versus the unfolded ones. For example, Fig. 4a shows the decay of the S(t) component μ=ν=x, with the x axis alligned in the direction connecting the carboxy and amino termini (which was also the direction of pull for cases when Fext was applied). Within the folded state and in the absence of temperature increases or applied forces, S(t) decays slowly as a power law in time; the native contacts provide the protein with the ability to store stress. Upon moderate heating or under low external forces, such that folding is maintained, relaxation is still slow, similar to that in the absence of heating or pulling. However, for temperatures T and external forces Fext large enough to induce unfolding, the system exhibits a significantly faster, exponential tail (Fig. 5a). Hence, when the native contacts are maintained, the slow, power law stress–stress time correlation decay accompanies the presence of force peaks in P(f) and Pnb(fnb).
being the ν and μ components of the non-bonding pair force and distance, respectively, between two atoms i and j. We gauged stress relaxation by the decay of the normalized stress–stress time correlation function S(t)=(C(t)−C(∞))/(C(0)−C(∞)), where C(t)=〈σμν(t)σμν(0)〉. The correlations S(t) in all directions did slow down in the folded state versus the unfolded ones. For example, Fig. 4a shows the decay of the S(t) component μ=ν=x, with the x axis alligned in the direction connecting the carboxy and amino termini (which was also the direction of pull for cases when Fext was applied). Within the folded state and in the absence of temperature increases or applied forces, S(t) decays slowly as a power law in time; the native contacts provide the protein with the ability to store stress. Upon moderate heating or under low external forces, such that folding is maintained, relaxation is still slow, similar to that in the absence of heating or pulling. However, for temperatures T and external forces Fext large enough to induce unfolding, the system exhibits a significantly faster, exponential tail (Fig. 5a). Hence, when the native contacts are maintained, the slow, power law stress–stress time correlation decay accompanies the presence of force peaks in P(f) and Pnb(fnb).
We also explored how the average stress itself varies as a function of the increase in T and Fext; this would measure the change in the mechanical response of the proteins as native contacts are disrupted. To this end, we computed the particle-normalized average stress 〈σxx〉/N (angle brackets denote time averaging) and its components, the bonded and non-bonded contributions to the stress virial (Fig. 4b). Although the total average of 〈σxx〉/N varied as −kBT, in accord with the generalized equipartition theorem, the bonded and non-bonded terms have opposite signs (cf. Fig. 5b). This implies that there is mutual frustration between bonding and non-bonding interactions; on the average, conformations inside a confining basin minima of the bonding potential energy are on barrier tops (destabilizing, non-minima) of the non-bonding potential. The fact that frustration increases only slightly, if ever, upon unfolding (cf. Fig. 5b and its inset) is consistent with the minimal frustration principle49, which states that naturally occurring proteins have evolutionarily designed sequences that fold efficiently to an ensemble of structures with hardly any traps arising from mismatch between energetic contributions. It will be of interest to analyse force and virial stress distributions, and relaxation in native sequences versus decoy proteins50 comparatively for a microscopic description of frustration.
Discussion
In summary, how akin are folded proteins to jammed grains? Our simulations of force distributions and the time scales for dynamical slowdown of stress relaxation for folded versus unfolded proteins indicate several similarities, when comparisons are made with the corresponding measures in the granular states. However, a more varied connection between these measures and structural or external properties as described above for grains point to a more nuanced picture. Lessons learned from the complexities of granular jamming warrant further investigations in proteins, particularly as interactions in proteins are more complex, of multiple types, and on multiple length and time scales.
For example, mechanical response in jammed granular matter is fundamentally different from that of ordinary solids. Jammed grains can make ‘fragile’ materials2 that may jam or unjam, depending on the direction of the applied forces. Proteins seem to share such unusual mechanical properties in the sense that an individual protein exhibits different resilience to unfolding under external pulling with atomic force microscopy or single-molecule tweezers, depending on the direction of the pull44,51. Novel lock-in force spectroscopies with improved resolution52 can directly observe reversible folding–refolding events and have even been used to measure the distributions of refolding forces at various temperatures53. Repeated on circular permutants (same sequence and fold, but different termini), they may be used to directly probe equilibrium forces on different protein atoms in further testing of the jamming signatures found in our study. Fragility may also be linked with the criteria for brittle response as quantified by Hyeon and Thirumalai54; a protein is brittle or soft depending on the displacement of the free-energy barrier along the unfolding reaction coordinate as a result of the application of external forces. As such, it will be of interest to study the correlation between the external force-induced change in shape of the jamming peaks of P(F) and the change in free-energy profiles along different unfolding directions. More generally, approaches presented herein may be used to develop techniques borrowed from soft matter theory to characterize protein folding as induced self-organization of stress-propagating paths or force chains across the protein tertiary structure.
Studying proteins with jamming measures may be important in the design of biomimetic polymers with responses to external mechanical stress that can be modulated by optical or thermal means, or controlled depending on the direction of stress application. Results derived herein suggest a route to designing novel resilient materials via an optimization of particle composition to yield specific interactions that augment the peak in the force distribution, or to rank the mechanical strength across various material compositions depending on the value of α. Gauging force distributions and stress relaxation may shed light in cases where proteins can jam during their response to external forces as part of their function in vivo; for example, in muscles, in the cytoskeleton, or on passage through cellular pores.
Methods
Computational details
The initial co-ordinates for the simulations were crystal structures of ubiquitin (PDB id 1UBQ55), titin (1TIT56) and calmodulin (1LJK57) obtained from the Protein Data Bank. Molecular dynamics simulations were performed by 1 fs time-step integration of Langevin dynamics with a friction co-efficient of 10 ps−1 using the CHARMM38 software package with a united-atom force field and the analytical continuum electrostatics58 implicit solvent model. In separate simulations, the three protein structures were equilibrated and run at T=300 K, then independent equilibration and production was repeated at increasing temperatures in the 300–700 K interval in steps of 100 K, to induce complete protein unfolding. The variation of their radius of gyration Rg measured the degree of protein unfolding as shown Fig. 1d. Similarly, external forces Fext of increasing magnitude were applied at the two ends (all at T=300 K) up to 1,000 pN in steps of 100 pN. The Rg variation under Fext for the three proteins is recorded in Fig. 1e. For a trajectory cut-off time of 100 ns, ubiquitin was stable until T=600 K. Under Fext at 300 K (20 ns), it unfolded above forces of 600 pN. For the same cut-offs, the corresponding values of titin are 600 K and 500 pN. Calmodulin, which has less mechanical strength, unfolded in our simulations at 400 K and at pulling forces above 200 pN. Note that these transition values are simply those at which unfolding is induced on the relatively fast time scale of the simulations, and are not supposed to directly match those measured in bulk experiments of stability. For all T-dependent studies, simulations were 100 ns long. The non-equilibrium pulling simulations were performed with constant tension Fext applied at the ends of the protein until unfolding occured (typical times ranged from 10 to 100 ns). For all simulations where unfolding occurred, only the equilibrated unfolded structures were recorded and included in the force distribution, the average stress and the stress–stress correlation analysis. The stress was measured along a fixed axis in the protein to avoid global rotational effects. Stresses were calculated along the vector that connected the first and last backbone atoms. The unfolding forces were applied in the opposite directions along this vector also, such as to increase the distance between the two atoms.
Error in the force distributions of three molecules in the folded and unfolded states was estimated by the s.d. from the mean. We computed the error bar of each P(F) function measured over the entire trajectory by calculating this function from ten different independent shorter trajectories. The equilibrium part of the production trajectory was divided into ten equal parts and we then binned the P(F) from each. The s.d. of these P(F) from their mean was computed, and the resulting error bars are reported in Supplementary Figs S3-S8.
Additional information
How to cite this article: Jose, P. P. & Andricioaei, I. Similarities between protein folding and granular jamming. Nat. Commun. 3:1161 doi: 10.1038/ncomms2177 (2012).
References
Jaeger H. M., Nagel S. R., Behringer R. P. Granular solids, liquids, and gases. Rev. Mod. Phys. 68: 1259–1273 (1996)
Cates M. E., Wittmer J. P., Bouchaud J. P., Claudin P. Jamming, force chains, and fragile matter. Phys. Rev. Lett. 81: 1841–1844 (1998)
Angell C. A. Formation of glasses from liquids and biopolymers. Science 267: 1924–1935 (1995)
Liu A. J., Nagel S. R. Jamming is not cool anymore. Nature 396: 21–22 (1998)
Coppersmith S. N., Liu C. H., Majumdar S., Narayan O., Witten T. A. Model for force fluctuations in bead packs. Phys. Rev. E 53: 4673–4686 (1996)
Trappe V., Prasad V., Cipelletti L., Segre P. N., Weitz D. A. Jamming phase diagram of attractive particles. Nature 411: 772–775 (2001)
O'Hern C. S., Langer S. A., Liu A. J., Nagel S. R. Force distributions near jamming and glass transition. Phys. Rev. Lett. 86: 111–114 (2001)
Lacevik N., Glotzer S. C. Dynamical heterogeneity and jamming in glass forming liquids. J. Phys. Chem. B 108: 19623–19633 (2004)
Ashwin S. S., Brumer Y., Reichman D. R., Sastry S. Relationship between mechanical and dynamical properties of glass forming liquids. J. Phys. Chem. B 108: 19703–19710 (2004)
Keys A. S., Abate A. R., Glotzer S. C., Durian D. J. Measurement of growing dynamic length scales and prediction of jamming transition in a granular material. Nat. Phys. 3: 260–264 (2007)
Lu K., Brodsky E. E., Kavahpour. P. A thermodynamic unification of jamming. Nat. Phys. 4: 404–407 (2008)
Mari R., Krzakala F., Kurchan J. Jamming versus glass transition. Phys. Rev. Lett. 103: 025701 (2009)
Elmatad Y. S., Chandler D., Garrahan J. P. Corresponding states of structural glass formers. J. Phys. Chem. B 113: 5563–5567 (2009)
Silbert L. E. Jamming of frictional spheres and random loose packing. Soft Matter 6: 2918–2924 (2010)
Snoeijer J. H., Vlugt T. J. H., van Hecke M., van Saarloos W. Force network ensemble: A new approach to static granular matter. Phys. Rev. Lett. 92: 054302 (2004)
Majmudar T. S., Behringer R. P. Contact force measurements and stress-induced anisotropy in granular materials. Nature 435: 1079–1082 (2005)
Bi D., Zhang J., Chakraborty B., Behringer R. P. Jamming by shear. Nature 480: 355–358 (2011)
Somfai E., van Hecke M., Ellenbroek W. G., Shundyak K., van Saarloos W. Critical and noncritical jamming of frictional grains. Phys. Rev. E 75: 020301 (2007)
Olsson P., Teitel S. Critical scaling of shear viscosity at the jamming transition. Phys. Rev. Lett. 99: 178001 (2007)
Vagberg D., Valdez-Balderas D., Moore M. A., Olsson P., Teitel. S. Finite-size scaling at the jamming transition: corrections to scaling and the correlation-length critical exponent. Phys. Rev. E 83: 030303(R) (2011)
Hatano T. Growing length and time scales in a suspension of athermal particles. Phys. Rev. E 79: 050301 (2009)
Heussinger C., Barrat J.-L. Jamming transition as probed by quasistatic shear flow. Phys. Rev. Lett. 102: 218303 (2009)
Head D. Critical scaling and aging in cooling systems near the jamming transition. Phys. Rev. Lett. 102: 138001 (2009)
Liu C. H. et al. Force fluctuations in bead packs. Science 269: 513–515 (1995)
Corwin E. I., Jaeger H. M., Nagel S. R. Structural signature of jamming in granular media. Nature 435: 1075–1078 (2005)
Howell D., Behringer R. P., Veje C. Stress fluctuations in a 2d granular Couette experiment: a continuous transition. Phys. Rev. Lett. 82: 5241–5244 (1999)
Makse H. A., Johnson D. L., Schwartz L. M. Packing of compressible granular materials. Phys. Rev. Lett. 84: 4160–4163 (2000)
Tighe B. P., Socolar J. E. S., Schaeffer D. G., Mitchener W. G., Huber M. L. Force distributions in a triangular lattice of rigid bars. Phys. Rev. E 72: 031306 (2005)
Dijksman J. A., Wortel G. H., van Dellen L. T. H., Dauchot O., van Hecke M. Jamming, yielding, and rheology of weakly vibrated granular media. Phys. Rev. Lett. 107: 108303 (2011)
Reis P. M., Ingale R. A., Shattuck M. D. Caging dynamics in a granular fluid. Phys. Rev. Lett. 98: 188301 (2007)
Reis P. M., Ingale R. A., Shattuck M. D. Forcing independent velocity distributions in an experimental granular fluid. Phys. Rev. E 75: 051311 (2007)
Anfinsen C. B., Scheraga H. A. Experimental and theoretical aspects of protein folding. Adv. Prot. Chem 29: 205–300 (1975)
Frauenfelder H., Sligar S. G., Wolynes. P. G. The energy landscapes and motions of proteins. Science 254: 1598–1603 (1991)
Onuchic J. N., Wolynes P. G. Theory of protein folding. Curr. Opin. Struct. Biol. 14: 70–75 (2004)
Huang S. H., Ratliff K. S., Schwartz M. P., Spenner J. M., Matouschek A. Mitochondria unfold precursor proteins by unraveling them from their N-termini. Nat. Struct. Biol. 6: 1132–1138 (1999)
Bustamante C., Macosko J. C., Wuite G. J. L. Grabbing the cat by the tail: manipulating molecules one by one. Nat. Rev. Mol. Cell Biol. 1: 130–136 (2000)
Brujic J., Hermans R. I., Walther Z. K. A., Fernandez J. M. Single-molecule force spectroscopy reveals signature of glassy dynamics in the energy landscape of ubiquitin. Nat. Phys 2: 282–286 (2006)
Brooks B. R., Bruccoleri R. E., Olafson B. D., States D. J., Swaminathan S., Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics. J. Comput. Chem. 4: 187–217 (1983)
Klimov D. K., Thirumalai D. Stretching single-domain proteins: phase diagram and kinetics of force-induced unfolding. Proc. Natl Acad. Sci. USA 96: 6166–6170 (1999)
Klimov D. K., Thirumalai D. Lattice model studies of force-induced unfolding of proteins. J. Phys. Chem. B 105: 6648–6654 (2001)
Mueth D. M., Jaeger H. M., Nagel S. R. Force distribution in a granular medium. Phys. Rev. E 57: 3164–3169 (1998)
Brujic J., Edwards S. F., Hopkinson I., Makse H. A. Measuring the distribution of interdroplet forces in a compressed emulsion system. Phys. A 327: 201–212 (2003)
Junker J. P., Ziegler F., Reif M. Ligand depended equilibrium fluctuations of single calmodulin molecules. Science 323: 633–637 (2009)
Brockwell D. J. et al. Pulling geometry defines the mechanical resistance of a beta-sheet protein. Nat. Struct. Biol. 10: 731–737 (2003)
Rief M., Gautel M., Oesterhelt F., Fernandez J. M., Gaub H. E. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science 276: 1109–1112 (1997)
Tighe B. P., Snoeijer J. H., Vlugt T. J. H., van Hecke M. The force network ensemble for granular packings. Soft Matter 6: 2908–D2917 (2010)
Ono I. K. et al. Effective temperatures of a driven system near jamming. Phys. Rev. Lett. 89: 095703 (2002)
Miller B., O’Hern C., Behringer R. P. Stress-fluctuations for continuously sheared granular materials. Phys. Rev. Lett. 77: 3110–3113 (1996)
Bryngelson J. D., Onuchic J. N., Socci N. D., Wolynes P. G. Funnels, pathways, and the energy landscape of protein-folding - A synthesis. Prot. Struct. Funct. Genet. 21: 167–195 (1995)
Samudrala R., Levitt. M. Decoys ‘R’ us: A database of incorrect conformations to improve protein structure prediction. Prot. Sci. 9: 1399–1401 (2000)
Wilcox A. J., Choy J., Bustamante C., Matouschek A. Effect of protein structure on mitochondrial import. Proc. Natl Acad. Sci. USA 102: 15435–15440 (2005)
Schlierf M., Berkemeier F., Rief M. Direct observation of active protein folding using lock-in force spectroscopy. Biophys. J. 93: 3989–3998 (2007)
Schlierf M., Rief M. Temperature softening of a protein in single-molecule experiments. J. Mol. Biol. 354: 497–503 (2005)
Hyeon C., Thirumalai D. Measuring the energy landscape roughness and the transition state location of biomolecules using single molecule mechanical unfolding experiments. J. Phys. Condens. Matter 19: 11301–11327 (2007)
Vijay-Kumar S., Bugg C. E., Cook W. J. Structure of ubiquitin refined at 1.8A resolution. J. Mol. Biol. 194: 531–544 (1987)
Improta S., Politou A. S., Pastore A. Immunoglobulin-like modules from titin I-band: extensible components of muscle elasticity. Structure 4: 323–337 (1996)
Ishida H., Nakashima K., Kumakiand Y., Nakata M., Hikichi K., Yazawa. M. The solution structure of apocalmodulin from Saccharomyces cerevisiae implies a mechanism for its unique Ca2+ binding property. Biochemistry 41: 15536–15542 (2002)
Schaefer M., Karplus. M. A comprehensive analytical treatment of continuum electrostatics. J. Phys. Chem. 100: 1578–1599 (1996)
Acknowledgements
IA acknowledges the generous support of the National Science Foundation Career Award programme (CHE-0918817).
Author information
Authors and Affiliations
Contributions
P.P.J. ran the computer simulation codes and collected data. P.P.J. and I.A. designed the simulations, analysed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures S1-S8 (PDF 185 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Jose, P., Andricioaei, I. Similarities between protein folding and granular jamming. Nat Commun 3, 1161 (2012). https://doi.org/10.1038/ncomms2177
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/ncomms2177
This article is cited by
-
Membrane Protein Structure, Function, and Dynamics: a Perspective from Experiments and Theory
The Journal of Membrane Biology (2015)
-
Folding and jamming
Nature Materials (2012)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.