Abstract
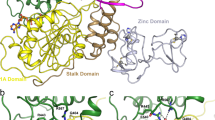
Here we report a highly conserved new binding site located at the interface between the protease and helicase domains of the hepatitis C virus (HCV) NS3 protein. Using a chemical lead, identified by fragment screening and structure-guided design, we demonstrate that this site has a regulatory function on the protease activity via an allosteric mechanism. We propose that compounds binding at this allosteric site inhibit the function of the NS3 protein by stabilizing an inactive conformation and thus represent a new class of direct-acting antiviral agents.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Reed, K.E. & Rice, C.M. Overview of hepatitis C virus genome structure, polyprotein processing and protein properties. Curr. Top. Microbiol. Immunol. 242, 55–84 (2000).
Bartenschlager, R. & Lohmann, V. Replication of hepatitis C virus. J. Gen. Virol. 81, 1631–1648 (2000).
Morgenstern, K.A. et al. Polynucleotide modulation of the protease, nucleoside triphosphatase, and helicase activities of the hepatitis C virus NS3–4A complex isolated from transfected COS cells. J. Virol. 71, 3767–3775 (1997).
Beran, R.K. & Pyle, A.M. Hepatitis C viral NS3–4a protease activity is enhanced by the NS3 helicase. J. Biol. Chem. 283, 29929–29937 (2008).
Beran, R.K., Serebrov, V. & Pyle, A.M. The serine protease domain of hepatitis C viral NS3 activates RNA helicase activity by promoting the binding of RNA substrate. J. Biol. Chem. 282, 34913–34920 (2007).
Rajagopal, V., Gurjar, M., Levin, M.K. & Patel, S.S. The protease domain increases the translocation stepping efficiency of the hepatitis C virus NS3–4A helicase. J. Biol. Chem. 285, 17821–17832 (2010).
Hopkins, A.L. et al. Complexes of HIV-1 reverse transcriptase with inhibitors of the HEPT series reveal conformational changes relevant to the design of potent non-nucleoside inhibitors. J. Med. Chem. 39, 1589–1600 (1996).
Kessl, J.J. et al. An allosteric mechanism for inhibiting HIV-1 integrase with a small molecule. Mol. Pharmacol. 76, 824–832 (2009).
Walker, M.A. New approaches for inhibiting HIV integrase: a journey beyond the active site. Curr. Opin. Investig. Drugs 10, 129–136 (2009).
Chen, K.X. & Njoroge, F.G. A review of HCV protease inhibitors. Curr. Opin. Investig. Drugs 10, 821–837 (2009).
Lamarre, D. et al. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426, 186–189 (2003).
Thibeault, D. et al. Use of the fused NS4a peptide-NS3 protease domain to study the importance of the helicase domain for protease inhibitor binding to hepatitis C virus NS3–4a. Biochemistry 48, 744–753 (2009).
Belon, C.A. & Frick, D.N. Helicase inhibitors as specifically targeted antiviral therapy for hepatitis C. Future Virol. 4, 277–293 (2009).
Frick, D.N. The hepatitis C virus NS3 protein: a model RNA helicase and potential drug target. Curr. Issues Mol. Biol. 9, 1–20 (2007).
Raney, K.D., Sharma, S.D., Moustafa, I.M., Cameron, C.E. & Hepatitis, C. Virus non-structural protein 3 (HCV NS3): a multifunctional antiviral target. J. Biol. Chem. 285, 22725–22731 (2010).
Chappell, K.J. et al. West Nile virus NS2B/NS3 protease as an antiviral target. Curr. Med. Chem. 15, 2771–2784 (2008).
Chernov, A.V. et al. The two-component NS2B–NS3 proteinase represses DNA unwinding activity of the West Nile virus NS3 helicase. J. Biol. Chem. 283, 17270–17278 (2008).
Murray, C.W. & Blundell, T. Structural biology in fragment-based drug design. Curr. Opin. Struct. Biol. 20, 497–507 (2010).
Murray, C.W. & Rees, D.C. The rise of fragment-based drug discovery. Nat. Chem. 1, 187–192 (2009).
Rhodes, D.I. et al. Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design. Antivir. Chem. Chemother. 21, 155–168 (2011).
Perryman, A.L. et al. Fragment-based screen against HIV protease. Chem. Biol. Drug Des. 75, 257–268 (2010).
Krimm, I., Lancelin, J.-M. & Praly, J.-P. Bindingevaluation of fragment-based scaffolds for probing allosteric enzymes. J. Med. Chem. 55, 1287–1295 (2012).
Maurer, T. et al. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc. Natl. Acad. Sci. USA 109, 5299–5304 (2012).
Pommier, Y. & Marchand, C. Interfacial inhibitors: targeting macromolecular complexes. Nat. Rev. Drug Discov. 11, 25–36 (2012).
Yao, N., Reichert, P., Taremi, S.S., Prosise, W.W. & Weber, P.C. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease–helicase. Structure 7, 1353–1363 (1999).
Lohmann, V. et al. Replication of subgenomic hepatitis C virus RNA in a hepatoma cell line. Science 285, 110–113 (1999).
Bartenschlager, R. Hepatitis C replicons: potential role for drug development. Nat. Rev. Drug Discov. 1, 911–916 (2002).
De Clercq, E. The design of drugs for HIV and HCV. Nat. Rev. Drug Discov. 6, 1001–1018 (2007).
Richman, D.D. The impact of drug resistance on the effectiveness of chemotherapy for chronic hepatitis B. Hepatology 32, 866–867 (2000).
Kuiken, C., Yusim, K., Boykin, L. & Richardson, R. The Los Alamos HCV sequence database. Bioinformatics 21, 379–384 (2005).
Ding, S.C., Kohlway, A.S. & Pyle, A.M. Unmasking the active helicase conformation of nonstructural protein 3 from hepatitis C virus. J. Virol. 85, 4343–4353 (2011).
Luo, D. et al. Flexibility between the protease and helicase domains of the Dengue virus NS3 protein conferred by the linker region and its functional implications. J. Biol. Chem. 285, 18817–18827 (2010).
Wang, W. et al. Conserved C-terminal threonine of hepatitis C virus NS3 regulates autoproteolysis and prevents product inhibition. J. Virol. 78, 700–709 (2004).
Wölk, B. et al. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3–NS4A complex expressed in tetracycline-regulated cell lines. J. Virol. 74, 2293–2304 (2000).
Brass, V. et al. Structural determinants for membrane association and dynamic organization of the hepatitis C virus NS3–4A complex. Proc. Natl. Acad. Sci. USA 105, 14545–14550 (2008).
Horner, S.M., Park, H.S. & Gale, M. Jr. Control of innate immune signaling and membrane targeting by the hepatitis C virus NS3/4A protease are governed by the NS3 helix α0. J. Virol. 86, 3112–3120 (2012).
Vonrhein, C. et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 67, 293–302 (2011).
Leslie, A.G.W. & Powell, H.R. Processing diffraction data with Mosflm. in Evolving Methods for Macromolecular Crystallography 245, 41–51 (2007).
Kabsch, W. Integration, scaling, spacegroup-assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 (2010).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Bricogne, G. et al. BUSTER version 1.11.2. (Global Phasing Ltd., 2011).
Mooij, W.T.M. et al. Automated protein-ligand crystallography for structure-based drug design. ChemMedChem 1, 827–838 (2006).
Acknowledgements
We acknowledge the contributions of M. Vinkovic, H. Angove, G.E. Besong, F. Holding, E. Chiarparin and M.L.Verdonk. We thank C. Cartwright for producing figures. X-ray data were collected at the European Synchrotron Radiation Facility and at the Diamond Lightsource. Sedimentation velocity experiments were performed by C. May and T. Laue at the Center to Advance Molecular Interaction Science at the University of New Hampshire. This research was supported by grant 087738 from the Wellcome Trust.
Author information
Authors and Affiliations
Contributions
S.M.S.-B. performed construct design and expression, protein purification, characterization and crystallization. G.C., A.J.W., M.G.C., S.D.H. and C.W.M. modeled and designed the compounds. A.J.W., M.G.C. and S.D.H. synthesized and characterized the compounds. J.C. performed ITC experiments. B.G. executed the western blot analysis and subgenomic replicon experiments and generated and characterized the resistance mutations. P.P. and P.A.W. crystallized the protein, collected the X-ray data and solved the crystal structures. S.J.R. and C.J.R. performed bioassay experiments. S.M.S.-B. and H.J. conceived the project. S.M.S.-B., A.J.W. and H.J. wrote the manuscript and managed the project. S.M.S-B., J.C., C.W.M. and H.J. conceived the model and designed the experiments.
Corresponding author
Ethics declarations
Competing interests
All authors are employees of Astex Pharmaceuticals and own shares in the company.
Supplementary information
Supplementary Text and Figures
Supplementary Methods and Supplementary Results (PDF 919 kb)
Rights and permissions
About this article
Cite this article
Saalau-Bethell, S., Woodhead, A., Chessari, G. et al. Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function. Nat Chem Biol 8, 920–925 (2012). https://doi.org/10.1038/nchembio.1081
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1081
This article is cited by
-
Natural HCV variants with increased replicative fitness due to NS3 helicase mutations in the C-terminal helix α18
Scientific Reports (2016)
-
Twenty years on: the impact of fragments on drug discovery
Nature Reviews Drug Discovery (2016)
-
Biophysics in drug discovery: impact, challenges and opportunities
Nature Reviews Drug Discovery (2016)
-
Functional interplay among the flavivirus NS3 protease, helicase, and cofactors
Virologica Sinica (2014)