Abstract
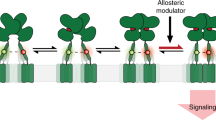
The controlled activation of proteins in living cells is an important goal in protein-design research, but to introduce an artificial activation switch into membrane proteins through rational design is a significant challenge because of the structural and functional complexity of such proteins. Here we report the allosteric activation of two types of membrane-bound neurotransmitter receptors, the ion-channel type and the G-protein-coupled glutamate receptors, using coordination chemistry in living cells. The high programmability of coordination chemistry enabled two His mutations, which act as an artificial allosteric site, to be semirationally incorporated in the vicinity of the ligand-binding pockets. Binding of Pd(2,2′-bipyridine) at the allosteric site enabled the active conformations of the glutamate receptors to be stabilized. Using this approach, we were able to activate selectively a mutant glutamate receptor in live neurons, which initiated a subsequent signal-transduction pathway.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Zorn, J. A. & Wells, J. A. Turning enzymes ON with small molecules. Nature Chem. Biol. 6, 179–188 (2010).
Rakhit, R., Navarro, R. & Wandless, T. J. Chemical biology strategies for posttranslational control of protein function. Chem. Biol. 21, 1238–1252 (2014).
Wolan, D. W., Zorn, J. A., Gray, D. C. & Wells, J. A. Small-molecule activators of a proenzyme. Science 326, 853–858 (2009).
Banaszynski, L. A., Chen, L.-C., Maynard-Smith, L. A., Ooi, A. G. L. & Wandless, T. J. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell 126, 995–1004 (2006).
Guo, Z., Zhou, D. & Schultz, P. G. Designing small-molecule switches for protein–protein interactions. Science 288, 2042–2045 (2000).
Qiao, Y., Molina, H., Pandey, A., Zhang, J. & Cole, P. A. Chemical rescue of a mutant enzyme in living cells. Science 311, 1293–1297 (2006).
Li, H., Hah, J.-M. & Lawrence, D. S. Light-mediated liberation of enzymatic activity: ‘small molecule’ caged protein equivalents. J. Am. Chem. Soc. 130, 10474–10475 (2008).
Arbely, E., Torres-Kolbus, J., Deiters, A. & Chin, J. W. Photocontrol of tyrosine phosphorylation in mammalian cells via genetic encoding of photocaged tyrosine. J. Am. Chem. Soc. 134, 11912–11915 (2012).
Li, J. et al. Palladium-triggered deprotection chemistry for protein activation in living cells. Nature Chem. 6, 352–361 (2014).
Li, J., Jia, S. & Chen, P. R. Diels–Alder reaction-triggered bioorthogonal protein decaging in living cells. Nature Chem. Biol. 10, 1003–1005 (2014).
Knight, Z. A. & Shokat, K. M. Chemical genetics: where genetics and pharmacology meet. Cell 128, 425–430 (2007).
Belshaw, P. J., Schoepfer, J. G., Liu, K.-Q., Morrison, K. L. & Schreiber, S. L. Rational design of orthogonal receptor–ligand combinations. Angew. Chem. Int. Ed. 34, 2129–2132 (1995).
Bishop, A. C. et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407, 395–401 (2000).
Baud, M. G. J. et al. A bump-and-hole approach to engineer controlled selectivity of BET bromodomain chemical probes. Science 346, 638–641 (2014).
Lu, Y., Yeung, N., Sieracki, N. & Marshall, N. M. Design of functional metalloproteins. Nature 460, 855–862 (2009).
Heinisch, T. & Ward, T. R. Design strategies for the creation of artificial metalloenzymes. Curr. Opin. Chem. Biol. 14, 184–199 (2010).
Yu, F. et al. Protein design: toward functional metalloenzymes. Chem. Rev. 114, 3495–3578 (2014).
Yeung, N. et al. Rational design of a structural and functional nitric oxide reductase. Nature 462, 1079–1082 (2009).
Faiella, M. et al. An artificial di-iron oxo-protein with phenol oxidase activity. Nature Chem. Biol. 5, 882–884 (2009).
Zastrow, M. L., Peacock, A. F. A., Stuckey, J. A. & Pecoraro, V. L. Hydrolytic catalysis and structural stabilization in a designed metalloprotein. Nature Chem. 4, 118–123 (2012).
Song, W. J. & Tezcan, F. A. A designed supramolecular protein assembly with in vivo enzymatic activity. Science 346, 1525–1528 (2014).
Elling, C. E., Thirstrup, K., Holst, B. & Schwartz, T. W. Conversion of agonist site to metal-ion chelator site in the β2-adrenergic receptor. Proc. Natl Acad. Sci. USA 96, 12322–12327 (1999).
Joh, N. H. et al. De novo design of a transmembrane Zn2+-transporting four-helix bundle. Science 346, 1520–1524 (2014).
Conklin, B. R. et al. Engineering GPCR signaling pathways with RASSLs. Nature Methods 5, 673–678 (2008).
Magnus, C. J. et al. Chemical and genetic engineering of selective ion channel–ligand interactions. Science 333, 1292–1296 (2011).
Goodey, N. M. & Benkovic, S. J. Allosteric regulation and catalysis emerge via a common route. Nature Chem. Biol. 4, 474–482 (2008).
De Smet, F., Christopoulos, A. & Carmeliet, P. Allosteric targeting of receptor tyrosine kinases. Nature Biotech. 32, 1113–1120 (2014).
Christopoulos, A. Allosteric binding sites on cell-surface receptors: novel targets for drug discovery. Nature Rev. Drug Discov. 1, 198–210 (2002).
Fehrentz, T., Schönberger, M. & Trauner, D. Optochemical genetics. Angew. Chem. Int. Ed. 50, 12156–12182 (2011).
Volgraf, M. et al. Allosteric control of an ionotropic glutamate receptor with an optical switch. Nature Chem. Biol. 2, 47–52 (2006).
Janovjak, H., Szobota, S., Wyart, C., Trauner, D. & Isacoff, E. Y. A light-gated, potassium-selective glutamate receptor for the optical inhibition of neuronal firing. Nature Neurosci. 13, 1027–1032 (2010).
Levitz, J. et al. Optical control of metabotropic glutamate receptors. Nature Neurosci. 16, 507–516 (2013).
Traynelis, S. F. et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol. Rev. 62, 405–496 (2010).
Fleming, J. J. & England, P. M. AMPA receptors and synaptic plasticity: a chemist's perspective. Nature Chem. Biol. 6, 89–97 (2010).
Armstrong, N. & Gouaux, E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron 28, 165–181 (2000).
Sobolevsky, A. I., Rosconi, M. P. & Gouaux, E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462, 745–756 (2009).
Armstrong, N., Jasti, J., Beich-Frandsen, M. & Gouaux, E. Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor. Cell 127, 85–97 (2006).
Holm, R. H., Kennepohl, P. & Solomon, E. I. Structural and functional aspects of metal sites in biology. Chem. Rev. 96, 2239–2314 (1996).
Kelso, M. J., Hoang, H. N., Appleton, T. G. & Fairlie, D. P. The first solution structure of a single α-helical turn. A pentapeptide α-helix stabilized by a metal clip. J. Am. Chem. Soc. 122, 10488–10489 (2000).
Hamachi, I. et al. Pd(en) as a sequence-selective molecular pinch for α-helical peptides. Chem. Lett. 30, 16–17 (2001).
Fujita, M., Tominaga, M., Hori, A. & Therrien, B. Coordination assemblies from a Pd(II)-cornered square complex. Acc. Chem. Res. 38, 369–378 (2005).
Pelton, J. G., Torchia, D. A., Meadow, N. D. & Roseman, S. Tautomeric states of the active-site histidines of phosphorylated and unphosphorylated IIIGlc, a signal-transducing protein from Escherichia coli, using two-dimensional heteronuclear NMR techniques. Protein Sci. 2, 543–558 (1993).
Miki, T. et al. LDAI-based chemical labeling of intact membrane proteins and its pulse-chase analysis under live cell conditions. Chem. Biol. 21, 1013–1022 (2014).
Niswender, C. M. & Conn, P. J. Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu. Rev. Pharmacol. Toxicol. 50, 295–322 (2010).
Rondard, P., Goudet, C., Kniazeff, J., Pin, J.-P. & Prézeau, L. The complexity of their activation mechanism opens new possibilities for the modulation of mGlu and GABAB class C G protein-coupled receptors. Neuropharmacology 60, 82–92 (2011).
Kunishima, N. et al. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature 407, 971–977 (2000).
Kobilka, B. K. & Deupi, X. Conformational complexity of G-protein-coupled receptors. Trends Pharmacol. Sci. 28, 397–406 (2007).
Deisseroth, K., Heist, E. K. & Tsien, R. W. Translocation of calmodulin to the nucleus supports CREB phosphorylation in hippocampal neurons. Nature 392, 198–202 (1998).
Benito, E. & Barco, A. CREB's control of intrinsic and synaptic plasticity: implications for CREB-dependent memory models. Trends Neurosci. 33, 230–240 (2010).
Chen, Y. et al. NS21: re-defined and modified supplement B27 for neuronal cultures. J. Neurosci. Methods 171, 239–247 (2008).
Acknowledgements
This work was funded by the Japan Science and Technology Agency CREST of Molecular Technologies and JSPS KAKENHI (JP15H01637) to I.H., Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research (B) to S.K. (25282238), Grants-in-Aid for Research Activity Start-up to R.K. (15H06318) and by the Ministry of Education, Culture, Sports, Science and Technology in Japan. We thank Y. Mori for providing the pEGFP-F plasmid and E. Gouaux for giving the plasmid of S1S2J. We also acknowledge the help of O. Hanpanich for constructing the plasmids of mGluR1.
Author information
Authors and Affiliations
Contributions
I.H. and S.K. conceived the project. R.K. and M.Y. conducted the construction of GluA2 plasmids. R.K. performed Ca2+ imaging in HEK293T and cultured cortical neurons, western blot analyses in cultured cortical neurons and the immunostaining experiment in HEK293T cells. R.K. conducted the construction of the plasmid of S1S2J(KR) and prepared the protein sample of S1S2J (WT and mutants) for 1H–15N HMQC NMR spectroscopy and fluorescence measurements. H.T. and M.S. performed and analysed 1H–15N HMQC NMR spectroscopy. S.K., R.K. and Y.M. performed the fluorescence measurements. Y.M. and M.Y. conducted the construction of the mGluR1 plasmids. Y.M. performed the fluorescent Ca2+ imaging in HEK293 cells. S.K. conducted the p-CREB assay in rat cortical neurons. T.N., R.I. and M.Y. contributed to the analysis and interpretation of Ca2+-imaging data. T.N. and R.I. conducted the electrophysiological measurements of cell capacitance and membrane potential. R.K., S.K. and I.H. wrote the manuscript. All the authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 2241 kb)
Rights and permissions
About this article
Cite this article
Kiyonaka, S., Kubota, R., Michibata, Y. et al. Allosteric activation of membrane-bound glutamate receptors using coordination chemistry within living cells. Nature Chem 8, 958–967 (2016). https://doi.org/10.1038/nchem.2554
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.2554
This article is cited by
-
Coordination chemogenetics for activation of GPCR-type glutamate receptors in brain tissue
Nature Communications (2022)
-
Organelle membrane-specific chemical labeling and dynamic imaging in living cells
Nature Chemical Biology (2020)