Abstract
The pharyngeal arch arteries (PAAs) are transient embryonic blood vessels that make indispensable contributions to the carotid arteries and great vessels of the heart, including the aorta and pulmonary arteries1,2. During embryogenesis, the PAAs appear in a craniocaudal sequence to connect pre-existing segments of the primitive circulation after de novo vasculogenic assembly from angioblast precursors3,4. Despite the unique spatiotemporal characteristics of PAA development, the embryonic origins of PAA angioblasts and the genetic factors regulating their emergence remain unknown. Here, we identify the embryonic source of PAA endothelium as nkx2.5+ progenitors in lateral plate mesoderm long considered to adopt cell fates within the heart exclusively5,6. Further, we report that PAA endothelial differentiation relies on Nkx2.5, a canonical cardiac transcription factor not previously implicated in blood vessel formation. Together, these studies reveal the heart field origin of PAA endothelium and attribute a new vasculogenic function to the cardiac transcription factor Nkx2.5 during great vessel precursor development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Congdon, E. D. Transformation of the aortic arch system during the development of the human embryo. Contrib. Embryol. 68, 49–110 (1922).
Moore, K. L. & Persaud, T. V. N. The Developing Human: Clinically Oriented Embryology 5th edn (Saunders, 1993).
Anderson, M. J., Pham, V. N., Vogel, A. M., Weinstein, B. M. & Roman, B. L. Loss of unc45a precipitates arteriovenous shunting in the aortic arches. Dev. Biol. 318, 258–267 (2008).
Li, P., Pashmforoush, M. & Sucov, H. M. Mesodermal retinoic acid signaling regulates endothelial cell coalescence in caudal pharyngeal arch artery vasculogenesis. Dev. Biol. 361, 116–124 (2012).
Ma, Q., Zhou, B. & Pu, W. T. Reassessment of Isl1 and Nkx2-5 cardiac fate maps using a Gata4-based reporter of Cre activity. Dev. Biol. 323, 98–104 (2008).
Schoenebeck, J. J., Keegan, B. R. & Yelon, D. Vessel and blood specification override cardiac potential in anterior mesoderm. Dev. Cell 13, 254–267 (2007).
Zhou, Y. et al. Latent TGF-beta binding protein 3 identifies a second heart field in zebrafish. Nature 474, 645–648 (2011).
Serbedzija, G. N., Chen, J. N. & Fishman, M. C. Regulation in the heart field of zebrafish. Development 125, 1095–1101 (1998).
Guner-Ataman, B. et al. Zebrafish second heart field development relies on progenitor specification in anterior lateral plate mesoderm and nkx2.5 function. Development 140, 1353–1363 (2013).
Hami, D., Grimes, A. C., Tsai, H. J. & Kirby, M. L. Zebrafish cardiac development requires a conserved secondary heart field. Development 138, 2389–2398 (2011).
Mosimann, C. et al. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development 138, 169–177 (2011).
Hiruma, T., Nakajima, Y. & Nakamura, H. Development of pharyngeal arch arteries in early mouse embryo. J. Anat. 201, 15–29 (2002).
Isogai, S., Horiguchi, M. & Weinstein, B. M. The vascular anatomy of the developing zebrafish: an atlas of embryonic and early larval development. Dev. Biol. 230, 278–301 (2001).
Stanley, E. G. et al. Efficient Cre-mediated deletion in cardiac progenitor cells conferred by a 3’UTR-ires-Cre allele of the homeobox gene Nkx2-5. Int. J. Dev. Biol. 46, 431–439 (2002).
Novak, A., Guo, C., Yang, W., Nagy, A. & Lobe, C. G. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis 28, 147–155 (2000).
Srinivas, S. et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 1478 (2001).
Kaufman, M. H. & Bard, J. B. L. The Anatomical Basis of Mouse Development 1st edn (Academic, 1999).
Targoff, K. L., Schell, T. & Yelon, D. Nkx genes regulate heart tube extension and exert differential effects on ventricular and atrial cell number. Dev. Biol. 322, 314–321 (2008).
Westerfield, M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) 4th edn (Univ. Oregon Press, 2000).
Nicoli, S. et al. MicroRNA-mediated integration of haemodynamics and Vegf signalling during angiogenesis. Nature 464, 1196–1200 (2010).
Sumanas, S. & Lin, S. Ets1-related protein is a key regulator of vasculogenesis in zebrafish. PLoS Biol. 4, 483–494 (2006).
Schilling, T. F. & Kimmel, C. B. Segment and cell type lineage restrictions during pharyngeal arch development in the zebrafish embryo. Development 120, 483–494 (1994).
Simoes, F. C., Peterkin, T. & Patient, R. Fgf differentially controls cross-antagonism between cardiac and haemangioblast regulators. Development 138, 3235–3245 (2011).
He, A., Kong, S. W., Ma, Q. & Pu, W. T. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc. Natl Acad. Sci. USA 108, 5632–5637 (2011).
Ferdous, A. et al. Nkx2-5 transactivates the Ets-related protein 71 gene and specifies an endothelial/endocardial fate in the developing embryo. Proc. Natl Acad. Sci. USA 106, 814–819 (2009).
Harris, I. S. & Black, B. L. Development of the endocardium. Pediatr. Cardiol. 31, 391–399 (2010).
Milgrom-Hoffman, M. et al. The heart endocardium is derivedfrom vascular endothelial progenitors. Development 138, 4777–4787 (2011).
McElhinney, D. B., Geiger, E., Blinder, J., Benson, D. W. & Goldmuntz, E. NKX2.5 mutations in patients with congenital heart disease. J. Am. College Cardiol. 42, 1650–1655 (2003).
Stoller, J. Z. & Epstein, J. A. Cardiac neural crest. Semin. Cell Dev. Biol. 16, 704–715 (2005).
Kikuchi, Y. et al. The zebrafish bonnie and clyde gene encodes a Mix family homeodomain protein that regulates the generation of endodermal precursors. Genes Dev. 14, 1279–1289 (2000).
Mizoguchi, T., Verkade, H., Heath, J. K., Kuroiwa, A. & Kikuchi, Y. Sdf1/Cxcr4 signaling controls the dorsal migration of endodermal cells during zebrafish gastrulation. Development 135, 2521–2529 (2008).
Jin, S. W., Beis, D., Mitchell, T., Chen, J. N. & Stainier, D. Y. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development 132, 5199–5209 (2005).
Traver, D. et al. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat. Immunol. 4, 1238–1246 (2003).
Yaniv, K. et al. Live imaging of lymphatic development in the zebrafish. Nat. Med. 12, 711–716 (2006).
Staal, J., Abramoff, M. D., Niemeijer, M., Viergever, M. A. & van Ginneken, B. Ridge-based vessel segmentation in color images of the retina. IEEE Trans. Med. Imag. 23, 501–509 (2004).
Burns, C. G. et al. High-throughput assay for small molecules that modulate zebrafish embryonic heart rate. Nat. Chem. Biol. 1, 263–264 (2005).
Long, S. & Rebagliati, M. Sensitive two-color whole-mount in situ hybridizations using digoxygenin- and dinitrophenol-labeled RNA probes. BioTechniques 32, 494 (2002) 496, 498 passim.
Thisse, C. & Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protocol. 3, 59–69 (2008).
Feng, H. et al. T-lymphoblastic lymphoma cells express high levels of BCL2, S1P1, and ICAM1, leading to a blockade of tumor cell intravasation. Cancer Cell 18, 353–366 (2010).
Marques, S. R., Lee, Y., Poss, K. D. & Yelon, D. Reiterative roles for FGF signaling in the establishment of size and proportion of the zebrafish heart. Dev. Biol. 321, 397–406 (2008).
Brand, M. G. & Nüsslein-Volhard, C. Keeping and Raising Zebrafish (Oxford Univ. Press, 2002).
Prall, O. W. et al. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell 128, 947–959 (2007).
Chapnik, E., Sasson, V., Blelloch, R. & Hornstein, E. Dgcr8 controls neural crest cells survival in cardiovascular development. Dev. Biol. 362, 50–56 (2012).
Lorandeau, C. G., Hakkinen, L. A. & Moore, C. S. Cardiovascular development and survival during gestation in the Ts65Dn mouse model for Down syndrome. Anat. Rec. 294, 93–101 (2011).
Acknowledgements
We are grateful to S. Paskaradevan and I. Scott (University of Toronto, USA) for their training in blastula transplantation. We thank C. Kinney for creating Supplementary Fig. 1; P. Obregon and T. Cashman for technical assistance in generating the Tg(nkx2.5:kaede) and Tg(nkx2.5:nZsYellow) transgenic lines, respectively; W. Goessling (Harvard Medical School, USA) for providing Tg(sox17:GFP) fish; T. North, and L. Zon (Harvard Medical School, USA) for providing Tg(kdrl:GFP) fish; I. Drummond (Massachusetts General Hospital, USA) for providing bonm425 fish; B. Barut (Harvard Medical School, USA) and L. Zon for providing bacterial artificial chromosomes; T. Evans (Weill Cornell Medical College, USA) for providing nkx2.5 plasmid for probe generation; G. Wilkinson (Medical College of Wisconsin, USA) and S. Sumanas (Cincinnati Children’s Hospital Medical Center, USA) for providing etsrp71 plasmid for probe generation; and the MGH Nephrology Division for access to their confocal microscopy facilities. We thank A. Vasilyev for assistance with confocal microscopy. N.P.L was supported by the Harvard Stem Cell Institute Training Grant (5HL087735) and a National Research Service Award (1F32HL 112579) from the National Heart, Lung and Blood Institute (NHLBI). B.G-A. was financially supported by an American Heart Association (AHA) Post-Doctoral Fellowship (10POST4170037). K.R.N. is financially supported by a National Research Service Award (5F32HL110627) from the National Heart, Lung and Blood Institute (NHLBI). R.P.H. is supported by grants from the National Health and Medical Research Council of Australia (NHMRC; 573732, 573703), Atlantic Philanthropies (19131) and National Heart Foundation of Australia (G08S3718). R.P.H. holds an NHMRC Australia Fellowship (573705). This work was financially supported by awards from the National Heart Lung and Blood Institute (5R01HL096816), American Heart Association (Grant in Aid no. 10GRNT4270021), and Harvard Stem Cell Institute (Seed Grant) to C.G.B. and the National Heart Lung and Blood Institute (5R01HL111179), the March of Dimes Foundation (FY12-467), and the Harvard Stem Cell Institute (Seed Grant and Young Investigator Award) to C.E.B. R.P.H. dedicates this work to Irene Myrtle Harvey, 1923–2013, from whom scholarship and inspiration were drawn.
Author information
Authors and Affiliations
Contributions
N.P-L. designed and performed the zebrafish experiments, analysed data and co-wrote the paper; R.S. designed, performed and analysed the mouse experiments, and co-wrote the paper; C.G.B. and B.G-A. created the Tg(nkx2.5:CreERT2) and Tg(nkx2.5:nZsYellow) lines; K.R.N., E.O.L. and L.J. performed and analysed zebrafish experiments; R.P.H. analysed data, designed experiments and co-wrote the paper; C.G.B. and C.E.B. initiated and directed the study, analysed data, and co-wrote the paper with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
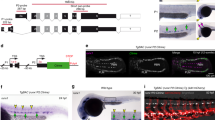
Supplementary Figure 1 Cartoons depicting nkx2.5+ heart field origin of PAA endothelium.
a, At 16 hpf (14ss), the bilateral nkx2.5+ ALPM (dark gray) contains progenitors for both cardiac and PAA endothelial lineages. b, As cardiac progenitors (red) migrate to the midline (18 hpf, 18ss) to form the primitive heart, nkx2.5+ PAA progenitors (light blue) remain lateral and begin to condense. c, As the heart tube elongates (30 hpf), nkx2.5+ PAA progenitors condense into 3 clusters with diffuse progenitors residing caudally. The anterior-most cluster (light gray) gives rise to non-PAA tissue within the face (data not shown). The 2nd cluster initiates expression of angioblast marker tie1 (green) and concomitantly downregulates nkx2.5 prior to forming PAA3. d, By 38 hpf, the 3rd cluster similarly adopts a tie1+ angioblast fate and subsequently forms PAA4. During similar stages, 2 undifferentiated caudal clusters emerge that are comprised of nkx2.5+ PAA progenitors specified in the heart field (light blue) and PAA progenitors initiating nkx2.5 expression de novo in the pharynx (dark blue). These caudal clusters form PAAs 5 and 6, respectively. e, By 60 hpf, PAAs 3-6 (green) are patent vessels expressing markers of terminal endothelial differentiation such as tie1, fli1a, and kdrl, but not the progenitor marker nkx2.5. f, Schematic diagram depicting the reciprocal relationship between nkx2.5 and tie1 expression during the PAA progenitor to angioblast transition. a-d, dorsal views, anterior up; e, lateral view, anterior left; angioblast cluster and corresponding PAA numbers indicated.
Supplementary Figure 2 ZsYellow is expressed in the anterior lateral plate and pharyngeal mesoderm of Tg(nkx2.5:ZsYellow) in patterns non-overlapping with hemangioblast or endodermal markers.
a, Gross evaluation of Tg(nkx2.5:ZsYellow) embryos at 4 dpf revealed robust ZsYellow fluorescence in the heart (H) and liver (L). b,c, ZsYellow transcripts (red) were detected in the anterior lateral plate mesoderm (ALPM), abutting hemangioblast markers etsrp71 (b) and scl (c). d,Tg(nkx2.5:ZsYellow); Tg(sox17:GFP) embryo immunostained with anti-ZsYellow and anti-GFP antibodies to highlight nkx2.5+ pharyngeal cells (red) and sox17+ pharyngeal endoderm (green), respectively. Red-only nkx2.5+ clusters (arrowheads) were detected in between green-only sox17+ endodermal pouches. e,f, ZsYellow+ populations were examined in endoderm-less bonnie and clyde (bon); Tg(nkx2.5:ZsYellow) embryos and their phenotypically wild-type (WT) siblings. A midline heart and bilateral pharyngeal clusters (arrowheads) were detected in WT siblings. bon mutants exhibited cardia bifida (asterisks) and significant populations of bilateral pharyngeal ZsYellow+ cells (brackets), demonstrating their non-endodermal nature. The un-clustered appearance of ZsYellow+ PAA progenitor cells in bon embryos likely reflects their lack of pharyngeal segmentation. WT, n = 78; bon, n = 25. g–p, Spatiotemporal comparison of endogenous nkx2. 5 and ZsYellow transcripts in wild-type and Tg(nkx2.5:ZsYellow) embryos. Transcripts for nkx2.5 andZsYellow were indistinguishable at 14ss (g,h), 18ss (i,j) and 28 hpf (k,l). While nkx2.5 transcripts disappeared specifically from pharyngeal mesoderm by 48 hpf (m,o), ZsYellow transcripts persisted in this region reflecting higher ZsYellow transcript stability (n,p). a,d,o,p lateral views, anterior left;b,c flat-mount dorsal views, anterior left; e-n dorsal views, anterior up; a-d, g-p n>20 embryos per group. Scale bar = 50 μm. Abbr: H, heart; L, liver.
Supplementary Figure 3 Fate mapping and genetic lineage tracing of nkx2.5+ cells.
a-i, nkx2.5+ pharyngeal clusters are derived from the heart field and give rise to PAA endothelium. a-c, Kaede photoconversion of cluster 3 (white box) at 30 hpf. Embryos were imaged in the green and red channels immediately after photoconversion (merged image is shown in a) and again at 60 hpf [green (inset, b), red (b) and merged images (c) are shown; n = 2]. Cluster 3 gave rise predominately to endothelium in PAA4 with a minimal contribution to PAA5 (b,c). d-f, When the left ALPM was photoconverted as shown in Figure 2k, the right ALPM (d) was left unconverted (white box) as a contralateral control. Embryos were imaged in the green and red channels immediately after photoconversion (merged image is shown in d) and subsequently at 60 hpf [green (e) and red (f) images are shown; (n = 2)]. No red reporter fluorescence was detected in the right side PAAs (asterisks in f). g-i, Tg(nkx2.5:Kaede) embryos were pan-photoconverted at 14ss and immediately imaged in the green and red channels (merged image is shown in g) and subsequently at 30 hpf [green (h) and red (i) images are shown; n = 3]. Red reporter fluorescence was detected in the heart tube (H) and three bilateral pairs of pharyngeal clusters (arrowheads, n = 3). j–t, Genetic lineage tracing of nkx2.5+ progenitors. j, Cartoon depicting vehicle control (EtOH; gray bar) or 4-HT (yellow bar) treatment of Tg(nkx2.5:ERCreT2);Tg(kdrl:CSY) or Tg(nkx2.5:ERCreT2); Tg(ubi:Switch) zebrafish embryos between tailbud and 8ss (10-13 hpf). Following extensive washing with fresh E3, embryos were incubated until 5 dpf and evaluated for reporter expression. k, Merged blue and yellow confocal z-stacks showing PAAs 3-6 in an EtOH-treated Tg(nkx2.5:ERCreT2); Tg(kdrl:BSY) embryo. Yellow reporter fluorescence was not detected (n = 80), across three experimental replicates. l, Merged green and red confocal z-stacks showing PAAs 3-6 in an EtOH-treated Tg(nkx2.5:ERCreT2); Tg(ubi:Switch) control embryo. Red reporter fluorescence was not detected (n = 80). m,n, Red (m) and merged (n, red and green) confocal z-stack images showing PAAs 3-6 in a 4-HT-treated Tg(nkx2.5:ERCreT2); Tg(ubi:Switch) embryo. Robust red reporter fluorescence was detected in PAAs 3-6 and the quantification is shown in o, n = 75 total across three experimental replicates. Error bars equal one standard deviation. p-t, Embryos derived from Nkx2−5IRESCre driver and ROSAYFP (q,r,s,t) or Z/EG (p) reporter line crosses co-stained with PECAM1 (red) and DAPI (blue). Embryos immunostained with anti-GFP (green) and anti-PECAM1 (red) antibodies showed contributions of Nkx2-5-lineage traced cells to the myocardium, endocardium, BAE and BMP (p,q, n = 2). r-t, Arrows indicate YFP+PECAM+ lineage traced endothelial cells. t, YFP+ lineage traced endothelial cells at the junction between the Aortic Sac (AS) and the left and right sides of PAA3. Cells counted across two embryos: E9.5 (PAA 1, n = 244), E10.5 (PAA 2, n = 216, PAA 3, n = 1357, PAA3 + aortic sac, n = 642). a,d,g-i dorsal views, anterior up; b,c,e,f,k-n lateral views, anterior left; p-t, coronal sections. Scale bar = 50 μm. Abbr: ss, somite stage; EtOH, ethanol; 4HT, 4-hydroxytamoxifen; dpf, days post-fertilization; A, atrium; AVC, atrio-ventricular canal; RV, right ventricle; LV, left ventricle; BAE branchial arch epithelium; BMP, branchial myogenic plate; OFT, outflow tract; AS, aortic sac; PAA number and left (L) and right (R) designations indicated.
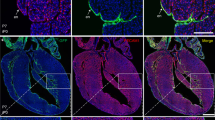
Supplementary Figure 4 nkx2.5 mutant and knock-down phenotypes in mice and zebrafish, respectively.
a-f, Validation of the nkx2.5 antisense morpholino.a, Schematic diagram of the nkx2.5 pre-mRNA and mature mRNA transcripts. The morpholino target site (MO, red bar) and the primer pairs used for RT-PCR (green arrows) are shown. b, Agarose gel of RT-PCR amplification products from control and nkx2.5 morphant (MOnkx2.5) 28 hpf embryos. While both control and nkx2. 5 morphant embryos contained pre-mRNA transcripts (primer pair F1R1, 189 bp amplicon, lanes 2 and 5), nkx2. 5 morphant embryos lacked spliced mRNA transcripts (primer pair F1R2, 230 bp amplicon, lanes 3 and 6). Lane 1 contains a 100 bp molecular weight ladder, and lanes 4 and 7 contain 18S rRNA loading controls. c-e, in situ hybridization analysis of tie1 transcripts revealed fewer PAA angioblast clusters in morphant (d, n = 65) when compared to control embryos (CTRL c, n = 33). The asterisk in (d) labels a single PAA angioblast. This phenotype was rescued by co-injection of 50 pg full-length zebrafish nkx2.5 mRNA (e, mRNAnkx2.5 n = 57). f, Phenotypic quantification and rescue; two-tailed t test, **P = 0.001, no significant (ns, P = 0.0755) difference was observed between CTRL and nkx2.5 morphants rescued with nkx2.5 mRNA. g-i, Section immunofluorescence analysis of Nkx2-5+/+ (g) and Nkx2-5lacZ/lacZ (h,i) embryos at E9.5 highlighting PECAM+ endothelium (red), alpha-actinin+ outflow tract (OFT) myocardium (green), and DAPI+ nuclei (blue) (n = 5 per group). Nkx2-5lacZ/lacZ embryos contain PECAM1+ endothelial cells in rudimentary PAA lumens (arrowhead, h) or scattered in regions where the dorsal aorta (DA) and 3rd PAA would otherwise form (arrows, i). d–e, Confocal z-stack images of flat-mounted control (j) and nkx2.5 morphant (MOnkx2.5; k) Tg(nkx2.5:nZsYellow) embryos at 14ss immunostained for ZsYellow protein (red). l, Graph depicting quantification of nkx2.5+ nuclei in the ALPM of control (n = 5) and MOnkx2.5 (n = 6) embryos. No significant (ns) difference was observed across two independent experiments. Error bars equal one standard deviation, two-tailed t test P = 0.1219. c-e lateral views, anterior up; g–i coronal sections; j,k flat-mounted dorsal views, anterior up. Scale bar = 50 μm. Abbr: OFT, outflow tract; DA, dorsal aorta.
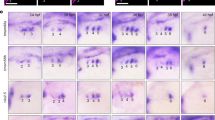
Supplementary Figure 5 Neither overexpression of nkx2.5 nor inhibition of FGF signaling suppresses PAA endothelial cell differentiation.
a-e, Embryos were injected with 100 pg full-length nkx2.5 mRNA (mRNAnkx2.5) and evaluated by in situ hybridization for scl expression in the ALPM at 7ss (a,b) or tie1 expression in pharyngeal clusters at 38 hpf (c,d). Expression of scl was severely reduced in the ALPM of nkx2.5 mRNA injected embryos (b) (n>20 per group). No significant (ns) difference (e) in the number of tie1-expressing PAA clusters was observed between control (c, n = 16) and nkx2.5 injected embryos (d, n = 26; two-tailed t test, P = 0.4481) across two experimental replicates. f,g Tg(cmlc2:EGFP) embryos were treated with DMSO or the FGF inhibitor SU5402 (10 μM) starting at 5ss and imaged at 48 hpf. Treated embryos exhibited a small ventricle, a phenotype known to arise from inhibition of FGF signaling. h-l, Tg(nkx2.5:ZsYellow) embryos were treated with DMSO and SU5402 (10 μM) beginning at the 5ss and imaged at 28 hpf (k,l) or fixed at 34 hpf for in situ hybridization (i,j). in situ hybridization for tie1 revealed equal numbers of PAA angioblast clusters (h) between control (i) and treated (j) embryos, (DMSO, n = 30; SU5402, n = 38; two-tailed ttest, P = 0.7254 across two experimental replicates). Interestingly, FGF-deficient embryos exhibited disrupted body vasculature as demonstrated by the absence of vessels in the eye and the LDA (bracket). ZsYellow+ clusters in the pharynx (arrowheads) were indistinguishable between control (k) and treated (l) animals, demonstrating effective segregation of the PAA and cardiac progenitors (n>20 per group). a,b,k,l dorsal views, anterior up; c,d,i,j lateral views, anterior left; f,g ventral views. Scale bar = 50 μm. Abbr: H, heart; LDA, lateral dorsal aorta; V, ventricle; A, atrium.
Supplementary information
Supplementary Information
Supplementary Information (PDF 669 kb)
Rights and permissions
About this article
Cite this article
Paffett-Lugassy, N., Singh, R., Nevis, K. et al. Heart field origin of great vessel precursors relies on nkx2.5-mediated vasculogenesis. Nat Cell Biol 15, 1362–1369 (2013). https://doi.org/10.1038/ncb2862
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2862
This article is cited by
-
Tmem88 confines ectodermal Wnt2bb signaling in pharyngeal arch artery progenitors for balancing cell cycle progression and cell fate decision
Nature Cardiovascular Research (2023)
-
Intrinsic myocardial defects underlie an Rbfox-deficient zebrafish model of hypoplastic left heart syndrome
Nature Communications (2022)
-
Exploring the Activities of RBPMS Proteins in Myocardial Biology
Pediatric Cardiology (2019)
-
LncRNA-uc.40 silence promotes P19 embryonic cells differentiation to cardiomyocyte via the PBX1 gene
In Vitro Cellular & Developmental Biology - Animal (2018)
-
Vascular heterogeneity and specialization in development and disease
Nature Reviews Molecular Cell Biology (2017)