Abstract
Interruption of combination antiretroviral therapy (ART) in HIV-1-infected individuals leads to rapid viral rebound. Here we report the results of a phase IIa open label clinical trial evaluating 3BNC117, a broad and potent neutralizing antibody (bNAb) against the CD4 binding site of HIV-1 Env1, in the setting of analytical treatment interruption (ATI) in 13 HIV-1-infected individuals. Participants with 3BNC117-sensitive virus outgrowth cultures were enrolled. Two or four 30 mg/kg infusions of 3BNC117, separated by 3 or 2 weeks, respectively, were generally well tolerated. The infusions were associated with a delay in viral rebound for 5-9 weeks after 2 infusions, and up to 19 weeks after 4 infusions, or an average of 6.7 and 9.9 weeks respectively, compared with 2.6 weeks for historical controls (p=<1e-5). Rebound viruses arose predominantly from a single provirus. In most individuals, emerging viruses showed increased resistance indicating escape. However, 30% of participants remained suppressed until antibody concentrations waned below 20 μg/ml, and the viruses emerging in all but one of these individuals showed no apparent resistance to 3BCN117, suggesting failure to escape over a period of 9-19 weeks. We conclude that administration of 3BNC117 exerts strong selective pressure on HIV-1 emerging from latent reservoirs during ATI in humans.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Scheid, J. F. et al. Sequence and Structural Convergence of Broad and Potent HIV Antibodies That Mimic CD4 Binding. Science, doi:science.1207227 [pii] 10.1126/science.1207227 (2011)
Scheid, J. F. et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458, 636–640, doi:nature07930 [pii] 10.1038/nature07930 (2009)
Klein, F. et al. Antibodies in HIV-1 vaccine development and therapy. Science 341, 1199–1204, doi:10.1126/science.1241144 (2013)
Barouch, D. H. et al. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature 503, 224–228, doi:10.1038/nature12744 (2013)
Halper-Stromberg, A. et al. Broadly neutralizing antibodies and viral inducers decrease rebound from HIV-1 latent reservoirs in humanized mice. Cell 158, 989–999, doi:10.1016/j.cell.2014.07.043 (2014)
Klein, F. et al. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature 492, 118–122, doi:10.1038/nature11604 (2012)
Horwitz, J. A. et al. HIV-1 suppression and durable control by combining single broadly neutralizing antibodies and antiretroviral drugs in humanized mice. Proceedings of the National Academy of Sciences of the United States of America 110, 16538–16543, doi:10.1073/pnas.1315295110 (2013)
Shingai, M. et al. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J Exp Med 211, 2061–2074, doi:10.1084/jem.20132494 (2014)
Caskey, M. et al. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 522, 487–491, doi:10.1038/nature14411 (2015)
Schoofs, T. et al. HIV-1 therapy with monoclonal antibody 3BNC117 elicits host immune responses against HIV-1. Science 352, 997–1001, doi:10.1126/science.aaf0972 (2016)
Lu, C. L. et al. Enhanced clearance of HIV-1-infected cells by broadly neutralizing antibodies against HIV-1 in vivo. Science 352, 1001–1004, doi:10.1126/science.aaf1279 (2016)
Kong, R. et al. Improving neutralization potency and breadth by combining broadly reactive HIV-1 antibodies targeting major neutralization epitopes. Journal of virology 89, 2659–2671, doi:10.1128/JVI.03136-14 (2015)
Lynch, R. M. et al. Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Science translational medicine 7, 319ra206, doi:10.1126/scitranslmed.aad5752 (2015)
Shingai, M. et al. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature 503, 277–280, doi:10.1038/nature12746 (2013)
Mouquet, H. et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proceedings of the National Academy of Sciences of the United States of America 109, E3268–3277, doi:10.1073/pnas.1217207109 (2012)
Rothenberger, M. K. et al. Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. Proceedings of the National Academy of Sciences of the United States of America 112, E1126–1134, doi:10.1073/pnas.1414926112 (2015)
Kearney, M. F. et al. Lack of detectable HIV-1 molecular evolution during suppressive antiretroviral therapy. PLoS pathogens 10, e1004010, doi:10.1371/journal.ppat.1004010 (2014)
Kearney, M. F. et al. Origin of Rebound Plasma HIV Includes Cells with Identical Proviruses that are Transcriptionally Active Before Stopping Antiretroviral Therapy. Journal of virology, 10.1128/JVI.02139-15 (2015)
Salantes, B. S. & B; Bar, Katharine. Potency and Kinetics of Autologous HIV-1 Neutralizing Antibody Responses During ATI. CROI Conference Abstracts Abstract #92 (2016)
West, A. P., Jr., Diskin, R., Nussenzweig, M. C. & Bjorkman, P. J. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proceedings of the National Academy of Sciences of the United States of America 109, E2083–2090, doi:10.1073/pnas.1208984109 (2012)
Diskin, R. et al. Restricting HIV-1 pathways for escape using rationally designed anti-HIV-1 antibodies. J Exp Med 210, 1235–1249, doi:10.1084/jem.20130221 (2013)
Lynch, R. M. et al. HIV-1 fitness cost associated with escape from the VRC01 class of CD4 binding site neutralizing antibodies. Journal of virology 89, 4201–4213, doi:10.1128/JVI.03608-14 (2015)
Zhou, T. et al. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity 39, 245–258, doi:10.1016/j.immuni.2013.04.012 (2013)
Lyumkis, D. et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342, 1484–1490, doi:10.1126/science.1245627 (2013)
Gautam, R. et al. A single injection of anti-HIV-1 antibodies protects against repeated SHIV challenges. Nature, 10.1038/nature17677 (2016)
Trkola, A. et al. Delay of HIV-1 rebound after cessation of antiretroviral therapy through passive transfer of human neutralizing antibodies. Nat Med 11, 615–622, doi:10.1038/nm1244 (2005)
Mehandru, S. et al. Adjunctive passive immunotherapy in human immunodeficiency virus type 1-infected individuals treated with antiviral therapy during acute and early infection. Journal of virology 81, 11016–11031, doi:10.1128/JVI.01340-07 (2007)
Ho, Y. C. et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155, 540–551, doi:10.1016/j.cell.2013.09.020 (2013)
Li, J. Z. et al. The size of the expressed HIV reservoir predicts timing of viral rebound after treatment interruption. AIDS 30, 343–353, doi:10.1097/QAD.0000000000000953 (2016)
West, A. P., Jr. Computational analysis of anti-HIV-1 antibody neutralization panel data to identify potential functional epitope residues. PNAS (2013)
Laird, G. M. et al. Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS pathogens 9, e1003398, doi:10.1371/journal.ppat.1003398 (2013)
Volberding, P. et al. Antiretroviral therapy in acute and recent HIV infection: a prospective multicenter stratified trial of intentionally interrupted treatment. AIDS 23, 1987–1995, doi:10.1097/QAD.0b013e32832eb285 (2009)
Kilby, J. M. et al. A randomized, partially blinded phase 2 trial of antiretroviral therapy, HIV-specific immunizations, and interleukin-2 cycles to promote efficient control of viral replication (ACTG A5024). The Journal of infectious diseases 194, 1672–1676, doi:10.1086/509508 (2006)
Jacobson, J. M. et al. Evidence that intermittent structured treatment interruption, but not immunization with ALVAC-HIV vCP1452, promotes host control of HIV replication: the results of AIDS Clinical Trials Group 5068. The Journal of infectious diseases 194, 623–632, doi:10.1086/506364 (2006)
Pereyra, F. et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330, 1551–1557, doi:10.1126/science.1195271 (2010)
Montefiori, D. C. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr Protoc Immunol Chapter 12 , Unit 12 11, doi:10.1002/0471142735.im1211s64 (2005)
Li, M. et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. Journal of virology 79, 10108–10125, doi:10.1128/JVI.79.16.10108-10125.2005 (2005)
Salazar-Gonzalez, J. F. et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. Journal of virology 82, 3952–3970, doi:10.1128/JVI.02660-07 (2008)
Kearse, M. et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649, doi:10.1093/bioinformatics/bts199 (2012)
Crooks, G. E., Hon, G., Chandonia, J. M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome research 14, 1188–1190, doi:10.1101/gr.849004 (2004)
Kirchherr, J. L. et al. High throughput functional analysis of HIV-1 env genes without cloning. Journal of virological methods 143, 104–111, doi:10.1016/j.jviromet.2007.02.015 (2007)
Xie, J. & Liu, C. Adjusted Kaplan-Meier estimator and log-rank test with inverse probability of treatment weighting for survival data. Statistics in medicine 24, 3089–3110, doi:10.1002/sim.2174 (2005)
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948, doi:btm404 [pii] 10.1093/bioinformatics/btm404 (2007)
Maddison, W. P. & Maddison, D. R. MacClade - Analysis of Phylogeny and Character Evolution - Version 4. (Sinauer Associates, Inc., 2001)
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. jModelTest 2: more models, new heuristics and parallel computing. Nature methods 9, 772, doi:10.1038/nmeth.2109 (2012)
Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic biology 59, 307–321, doi:10.1093/sysbio/syq010 (2010)
Chun, T. W. et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387, 183–188, doi:10.1038/387183a0 (1997)
Parrish, N. F. et al. Phenotypic properties of transmitted founder HIV-1. Proceedings of the National Academy of Sciences of the United States of America 110, 6626–6633, doi:10.1073/pnas.1304288110 (2013)
Maydt, J. & Lengauer, T. Recco: recombination analysis using cost optimization. Bioinformatics 22, 1064–1071, doi:10.1093/bioinformatics/btl057 (2006)
Giorgi, E. E. & Bhattacharya, T. A note on two-sample tests for comparing intra-individual genetic sequence diversity between populations. Biometrics 68, 1323–1326; author reply 1326, doi:10.1111/j.1541-0420.2012.01775.x (2012)
Altfeld, M. A. et al. Identification of dominant optimal HLA-B60- and HLA-B61-restricted cytotoxic T-lymphocyte (CTL) epitopes: rapid characterization of CTL responses by enzyme-linked immunospot assay. Journal of virology 74, 8541–8549 (2000)
Addo, M. M. et al. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. Journal of virology 77, 2081–2092 (2003)
Acknowledgements
We would like to thank the trial participants for their invaluable support; We thank the Rockefeller University Hospital Clinical Research Support Office and nursing staff for help with recruitment and study implementation, especially Noreen Buckley, Arlene Hurley, Sivan Ben Avraham Shulman and Lauren Corregano. All members of the Nussenzweig lab, especially Till Schoofs, Ari Halper-Stromberg, Mila and Zoran Jankovic. Cecille Unson-O'Brien, Juan Dizon, Renise Baptiste and Rebeka Levin for sample processing and study coordination; Audrey Louie for regulatory support; Pat Fast and Harriet Park for clinical monitoring. Elena Giorgi and William Fischer from Los Alamos National Laboratory. Rajesh Tim Gandhi, Jonathan Li and The AIDS Clinical Trials Group (grant UM1 AI068636) and its Statistical and Data Management Center (grant UM1 AI068634). This study was supported by the following grants: Collaboration for AIDS Vaccine Discovery grant OPP1033115 (M.C.N.) and OPP1032144 (M.S.S.). Grant 8 UL1 TR000043 from the National Center for Advancing Translational Sciences (NCATS); NIH Clinical and Translational Science Award (CTSA) program; NIH Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (CHAVI-ID) 1UM1 AI100663-01 (M.C.N) and 5UM1 AI100645-03 (B.H.H.); Bill and Melinda Gates Foundation grants OPP1092074 and OPP1124068 (M.C.N); NIH HIVRAD P01 AI100148 (PJB and MCN); the Robertson Foundation to M.C.N. M.C.N. is a Howard Hughes Medical Institute Investigator. Ruth L. Kirschstein National Research Service Award. F30 AI112426 (E.F.K); F31 AI118555 (J.A.H.); The NIH Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (CHAVI-ID) 1UM1 AI00645 (B.H.H.); The University of Pennsylvania Center for AIDS Research (CFAR) Single Genome Amplification Service Center P30 AI045008 (B.H.H.); The NIH Scripps Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (CHAVI-ID and 1UM1-AI100663) (B.D.W).
Author information
Authors and Affiliations
Contributions
M.C.N, J.F.S, J.A.H and M.C wrote the manuscript; J.F.S, M.C and M.C.N designed the trial; J.F.S, J.A.H, Y.B, J.C.C.L, L.N, Y.Z.C, C-L.L and M.B performed tissue culture experiments and SGS amplifications; M.S.S performed TZM-bl assays; J.F.S, J.A.H, Y.B, E.F.K, T.O, A.P.W, G.H.L, P.J.B, F.K, S.J.S, B.H.H, M.C.N and M.C analyzed the data; E.F.K, G.H.L and B.H.H performed SGA analysis; I.S, R.P and J.F.S processed patient samples; L.B, S.H, A.S, M.W-P, B.Z, R.M.G, S.J.S and M.C performed patient recruitment; A.F and N.P performed statistical analyses; B.J and B.D.W performed antigen specific T cell experiments; T.K and T.H produced 3BNC117 and provided PK data.
Corresponding authors
Ethics declarations
Competing interests
There are pending patent applications on the 3BNC117 and 10-1074 antibodies by Rockefeller University on which M.C.N. and J.F.S. are inventors. The patents are not licensed by any companies. Neither M.C.N. nor J.F.S. have any competing financial interests.
Additional information
There are pending patent applications on the 3BNC117 and 10-1074 antibodies by Rockefeller University on which M.C.N and J.F.S are inventors. The patents are not licensed by any companies.
Reviewer Information Nature thanks S. Deeks, D. Richman and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
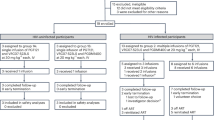
Extended Data Figure 1 Study Participant Selection and Neutralization of Pre-Infusion Cultures by 3BNC117.
a, Flow diagram showing selection of study participants. b, Bar diagrams showing IC50 values (μg/ml) in TZM-bl assays for 3BNC117 against bulk virus outgrowth culture supernatants from the indicated time point pre-infusion for each participant (Supplementary Table 3). In some participants both screen and day 0 cultures were obtained and showed a less than three-fold variation in IC50 values. The red dotted line indicates an IC50 of 2 μg/ml which was used as a threshold for inclusion in the study.
Extended Data Figure 2 CD4+ and CD8+ T cells during Study Period in Participants.
Absolute T cell counts (a, c) and percent CD4+ and CD8+ T cells among CD3+T cells (b, d) for Group A and B respectively (Supplementary Table 4). 3BNC117 infusions are indicated with red arrows. e, Comparison of absolute CD4+ T cell counts and percent CD4+ T cells among CD3+T cells at screen, day 0, rebound and after re-suppression. Shown is the data from participants 701, 702, 703, 704, 707, 708, 709, 711 and 713 for whom re-suppression CD4 counts were available (Supplementary Table 4). The last available time point was used as re-suppression time point. Red lines indicate the mean value and error bars indicate standard deviation. p-values were obtained using a paired t-test comparing the indicated time points. f, Plasma viral loads and CD4 counts in all study participants. 3BNC117 infusions are indicated with red arrows. The left y-axis shows plasma viral loads in RNA copies/ml (black curves), and right y-axis shows absolute CD4 counts in cells/μl (red curves). Gray areas indicate ART therapy.
Extended Data Figure 3 HIV-specific T-cell responses.
Total breadth (open squares) and magnitude (bars) of T-cell responses against HIV-1 overlapping peptides at the designated time points following administration of 3BNC117 (yellow arrows indicate infusions of 3BNC117 at 30 mg/kg). For all study participants, antiretroviral therapy was discontinued on day 2 after the first 3BNC117 administration. Blue arrows indicate the time of viral rebound. For study participants 710, 712 and 715 rebound occurred at week 19, 16 and 11, respectively. Baseline samples for study participant 710 and week 12 samples for study participant 714 were not available for ELISpot analysis. Overall, breadth, magnitude and protein specificity were heterogeneous among the study participants.
Extended Data Figure 4 Viral Rebound in ACTG Control Subjects and Trial Participants.
a, Kaplan-Meier plot summarizing viral rebound in 52 ACTG trial participants who underwent ATI without antibody treatment (black, Supplementary Table 6) and trial participants (Fig. 2 a, b, Supplementary Table 4). Six Group A participants are shown in red, seven Group B participants in blue and the combination in green as indicated. y-axis indicates % participants with viral levels below 200 RNA copies/ml, x-axis indicates weeks after ATI initiation. The survival curves of all considered partitions of the trial participants (Group A, Group B and Group A + B) differed significantly at significance level α = 0.05 from the survival curve of the ACTG trial participants. For the comparison of Group A (Group A + B) with the ACTG trial participants, we performed a weighted log-rank test adjusting for the clinical variables ‘years on ART’ and ‘age’ to correct for possible confounding factors (Supplementary Table 7, p-value < 0.00001). We identified those potential confounders by univariate parametric survival regression using a likelihood ratio test (Statistical Methods). Since we did not discover any confounders with the same analysis among all available clinical variables for the comparison between Group B participants and the ACTG trial participants, we performed a standard log-rank test in that setting (p-value < 0.0001). b-d, In order to perform a survival regression, the distribution of the rebound times has to be determined. Therefore, we compared the empirical cumulative distribution function (CDF) of the rebound times (black, solid line) with the CDF of the rebound times to a fitted distribution (Weibull, exponential, normal, logistic, log-normal, and gamma) for each comparison group (combined trial participants, group A or group B with ACTG control patients). Since the Akaike Information Criterion (AIC) and the Bayesian Information Criterion (BIC) were smallest for the log-normal distribution (green), we have chosen to model the rebound times with the log-normal distribution. e, Dot plot indicating the relationship between cell associated HIV DNA in pre-infusion PBMCs (y-axis) and the week of rebound (x-axis). Group A and group B participants are colored red and blue respectively. The p-value was derived from calculating the pearson correlation coefficient.
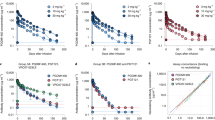
Extended Data Figure 5 In vitro neutralization of pre-infusion and rebound virus outgrowth cultures by 3BNC117 or 10-1074.
TZM-bl assay neutralization by 3BNC117 (a) and 10-1074 (b) are shown for individual virus outgrowth cultures derived from pre-infusion (black lines/symbols) or rebound (red lines/symbols) time points for each participant. In some cases, multiple independent cultures were grown from a single time point and assayed for neutralization (Supplementary Table 3). “Screen” refers to cultures of PBMC samples taken weeks before infusion during screening, while “Day 0” refers to cultures of PBMC collected immediately before the first 3BNC117 infusion. Rebound culture time points are denoted by the week (“W”) at which the samples were collected. Symbols reflect the means of two technical replicates; error bars denote standard deviation.
Extended Data Figure 6 Phylogenetic tree of env nucleotide sequences from trial participants.
A maximum likelihood phylogenetic tree was constructed from single genome derived viral env sequences from outgrowth culture supernatants as well as plasma from participants 701 (olive), 702 (black), 703 (pink), 704 (yellow), 707 (light blue), 708 (green), 709 (dark blue) and 711 (brown). Hypervariable (as defined in http://www.hiv.lanl.gov/content/sequence/VAR_REG_CHAR/) and other poorly aligned regions were excluded from the analysis. The tree was constructed using PhyML with a GTR+I+G substitution model and midpoint rooted. Asterisks indicate 100% bootstrap support (only values for major nodes are shown). The scale bar indicates 0.01 substitutions per site.
Extended Data Figure 7 Rebound virus clonality and neutralization sensitivity to 3BNC117.
Maximum likelihood phylogenetic trees of plasma and culture-derived env sequences are shown for participants 701, 702, 703, 704. Sequences obtained at screening, on Day 0, and consecutive rebound time points (plasma and cultures) are color coded as indicated. The trees were rooted based on the branch insertion identified in the between-subject tree (Extended Data Fig. 6). Bootstrap values ≥90% are shown. Names of env sequences used to generate pseudoviruses for 3BNC117 neutralization analysis are indicated along with the respective IC80 titers in μg/ml. Representative rebound viruses selected in Fig. 4b are marked with red stars (Fig. 4b, Supplementary Table 9). Zero branch length viruses in multi rebounders 702 and 703 are marked with black stars.
Extended Data Figure 8 Rebound virus clonality and neutralization sensitivity to 3BNC117.
Maximum likelihood phylogenetic trees of plasma and culture-derived env sequences are shown for participants 707, 708, 709 and 711. Sequences obtained at screening, on Day 0, and consecutive rebound time points (plasma and cultures) are color coded as indicated. The trees were rooted based on the branch insertion identified in the between-subject tree (Extended Data Fig. 6). Bootstrap values ≥90% are shown. Names of env sequences used to generate pseudoviruses for 3BNC117 neutralization analysis are indicated along with the respective IC80 titers in μg/ml. Representative rebound viruses selected in Fig. 4b are marked with red stars (Fig. 4b, Supplementary Table 9). Zero branch length viruses in multi rebounder 709 are marked with black stars.
Supplementary information
Supplementary Information
This file contains supplementary Figures 1-2, Supplementary Tables 1-9 and Supplementary References. This file was updated on 27 July 2016 to correct the reference list. (PDF 11137 kb)
Rights and permissions
About this article
Cite this article
Scheid, J., Horwitz, J., Bar-On, Y. et al. HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption. Nature 535, 556–560 (2016). https://doi.org/10.1038/nature18929
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature18929