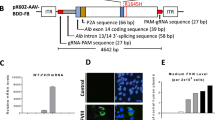
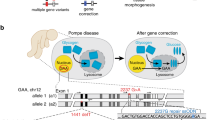
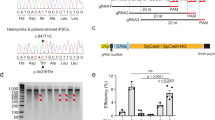
Abstract
Editing of the human genome to correct disease-causing mutations is a promising approach for the treatment of genetic disorders. Genome editing improves on simple gene-replacement strategies by effecting in situ correction of a mutant gene, thus restoring normal gene function under the control of endogenous regulatory elements and reducing risks associated with random insertion into the genome. Gene-specific targeting has historically been limited to mouse embryonic stem cells. The development of zinc finger nucleases (ZFNs) has permitted efficient genome editing in transformed and primary cells that were previously thought to be intractable to such genetic manipulation1. In vitro, ZFNs have been shown to promote efficient genome editing via homology-directed repair by inducing a site-specific double-strand break (DSB) at a target locus2,3,4, but it is unclear whether ZFNs can induce DSBs and stimulate genome editing at a clinically meaningful level in vivo. Here we show that ZFNs are able to induce DSBs efficiently when delivered directly to mouse liver and that, when co-delivered with an appropriately designed gene-targeting vector, they can stimulate gene replacement through both homology-directed and homology-independent targeted gene insertion at the ZFN-specified locus. The level of gene targeting achieved was sufficient to correct the prolonged clotting times in a mouse model of haemophilia B, and remained persistent after induced liver regeneration. Thus, ZFN-driven gene correction can be achieved in vivo, raising the possibility of genome editing as a viable strategy for the treatment of genetic disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Urnov, F. D., Rebar, E. J., Holmes, M. C., Zhang, H. S. & Gregory, P. D. Genome editing with engineered zinc finger nucleases. Nature Rev. Genet. 11, 636–646 (2010)
Urnov, F. D. et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435, 646–651 (2005)
Porteus, M. H. & Baltimore, D. Chimeric nucleases stimulate gene targeting in human cells. Science 300, 763 (2003)
Bibikova, M. et al. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol. Cell. Biol. 21, 289–297 (2001)
Cartier, N. et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science 326, 818–823 (2009)
Aiuti, A. et al. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N. Engl. J. Med. 360, 447–458 (2009)
Cideciyan, A. V. et al. Human gene therapy for RPE65 isomerase deficiency activates the retinoid cycle of vision but with slow rod kinetics. Proc. Natl Acad. Sci. USA 105, 15112–15117 (2008)
Maguire, A. M. et al. Safety and efficacy of gene transfer for Leber’s congenital amaurosis. N. Engl. J. Med. 358, 2240–2248 (2008)
Bainbridge, J. W. et al. Effect of gene therapy on visual function in Leber’s congenital amaurosis. N. Engl. J. Med. 358, 2231–2239 (2008)
Cavazzana-Calvo, M. et al. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature 467, 318–322 (2010)
Howe, S. J. et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Invest. 118, 3143–3150 (2008)
Hacein-Bey-Abina, S. et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302, 415–419 (2003)
Hanna, J. et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science 318, 1920–1923 (2007)
Pollak, E. S. & High, K. A. in The Metabolic and Molecular Bases of Inherited Disease (eds Scriver, C. R., Beaudet, A. L., Valle, D. & Sly, W. S. ) 4393–4413 (McGraw-Hill, 2001)
Green, P. Haemophilia B Mutation Database 〈http://www.kcl.ac.uk/ip/petergreen/haemBdatabase.html〉 (2004)
Moehle, E. A. et al. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc. Natl Acad. Sci. USA 104, 3055–3060 (2007)
Manno, C. S. et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nature Med. 12, 342–347 (2006)
Miao, C. H. et al. Inclusion of the hepatic locus control region, an intron, and untranslated region increases and stabilizes hepatic factor IX gene expression in vivo but not in vitro . Mol. Ther. 1, 522–532 (2000)
Thompson, A. R. & Chen, S. H. Germ line origins of de novo mutations in hemophilia B families. Hum. Genet. 94, 299–302 (1994)
Zambrowicz, B. P. et al. Disruption of overlapping transcripts in the ROSA βgeo 26 gene trap strain leads to widespread expression of β-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl Acad. Sci. USA 94, 3789–3794 (1997)
Lin, H. F., Maeda, N., Smithies, O., Straight, D. L. & Stafford, D. W. A coagulation factor IX-deficient mouse model for human hemophilia B. Blood 90, 3962–3966 (1997)
Perez, E. E. et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nature Biotechnol. 26, 808–816 (2008)
Nakai, H. et al. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo . J. Virol. 75, 6969–6976 (2001)
Li, H. et al. Assessing the potential for AAV vector genotoxicity in a murine model. Blood 117, 3311–3319 (2011)
Nakai, H. et al. AAV serotype 2 vectors preferentially integrate into active genes in mice. Nature Genet. 34, 297–302 (2003)
Miller, D. G., Petek, L. M. & Russell, D. W. Adeno-associated virus vectors integrate at chromosome breakage sites. Nature Genet. 36, 767–773 (2004)
Doyon, Y. et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nature Biotechnol. 26, 702–708 (2008)
Ayuso, E. et al. High AAV vector purity results in serotype- and tissue-independent enhancement of transduction efficiency. Gene Ther. 17, 503–510 (2010)
Mitchell, C. & Willenbring, H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nature Protocols 3, 1167–1170 (2008)
Berry, C., Hannenhalli, S., Leipzig, J. & Bushman, F. D. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput. Biol. 2, e157 (2006)
Acknowledgements
This work was funded by the National Institutes of Health and the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
H.L., V.H., Y.D., T.L., S.L.M., P.D.G., M.C.H. and K.A.H. designed the experiments. H.L., V.H., Y.D., T.L., S.Y.W., A.S.B., N.M., X.M.A., R.S., L.I., S.L.M., J.D.F., F.R.K., S.Z., D.E.P. and E.J.R. generated reagents and performed the experiments. H.L., Y.D., F.D.B., P.D.G., M.C.H. and K.A.H. wrote and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
Y.D., T.L., S.Y.W., D.E.P., E.J.R., P.D.G. and M.C.H. were all employees of Sangamo Biosciences when involved in this work. K.A.H. holds patents related to AAV vectors in the treatment of haemophilia.
Supplementary information
Supplementary Figures
The file contains Supplementary Figures 1-14 with legends. (PDF 2722 kb)
Rights and permissions
About this article
Cite this article
Li, H., Haurigot, V., Doyon, Y. et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature 475, 217–221 (2011). https://doi.org/10.1038/nature10177
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10177
This article is cited by
-
Adenine transversion editors enable precise, efficient A•T-to-C•G base editing in mammalian cells and embryos
Nature Biotechnology (2024)
-
Methods for CRISPR-Cas as Ribonucleoprotein Complex Delivery In Vivo
Molecular Biotechnology (2023)
-
Therapeutic homology-independent targeted integration in retina and liver
Nature Communications (2022)
-
Application of Gene Therapy in Hemophilia
Current Medical Science (2022)
-
Ex vivo editing of human hematopoietic stem cells for erythroid expression of therapeutic proteins
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.