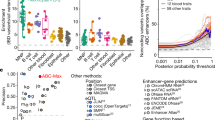
Abstract
Combined analyses of gene networks and DNA sequence variation can provide new insights into the aetiology of common diseases that may not be apparent from genome-wide association studies alone. Recent advances in rat genomics are facilitating systems-genetics approaches1,2. Here we report the use of integrated genome-wide approaches across seven rat tissues to identify gene networks and the loci underlying their regulation. We defined an interferon regulatory factor 7 (IRF73)-driven inflammatory network (IDIN) enriched for viral response genes, which represents a molecular biomarker for macrophages and which was regulated in multiple tissues by a locus on rat chromosome 15q25. We show that Epstein–Barr virus induced gene 2 (Ebi2, also known as Gpr183), which lies at this locus and controls B lymphocyte migration4,5, is expressed in macrophages and regulates the IDIN. The human orthologous locus on chromosome 13q32 controlled the human equivalent of the IDIN, which was conserved in monocytes. IDIN genes were more likely to associate with susceptibility to type 1 diabetes (T1D)—a macrophage-associated autoimmune disease—than randomly selected immune response genes (P = 8.85 × 10−6). The human locus controlling the IDIN was associated with the risk of T1D at single nucleotide polymorphism rs9585056 (P = 7.0 × 10−10; odds ratio, 1.15), which was one of five single nucleotide polymorphisms in this region associated with EBI2 (GPR183) expression. These data implicate IRF7 network genes and their regulatory locus in the pathogenesis of T1D.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Hubner, N. et al. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nature Genet. 37, 243–253 (2005)
Petretto, E. et al. Integrated genomic approaches implicate osteoglycin (Ogn) in the regulation of left ventricular mass. Nature Genet. 40, 546–552 (2008)
Honda, K. et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434, 772–777 (2005)
Pereira, J. P., Kelly, L. M., Xu, Y. & Cyster, J. G. EBI2 mediates B cell segregation between the outer and centre follicle. Nature 460, 1122–1126 (2009)
Gatto, D., Paus, D., Basten, A., Mackay, C. R. & Brink, R. Guidance of B cells by the orphan G protein-coupled receptor EBI2 shapes humoral immune responses. Immunity 31, 259–269 (2009)
Altshuler, D., Daly, M. J. & Lander, E. S. Genetic mapping in human disease. Science 322, 881–888 (2008)
Schadt, E. E. Molecular networks as sensors and drivers of common human diseases. Nature 461, 218–223 (2009)
Chen, Y. et al. Variations in DNA elucidate molecular networks that cause disease. Nature 452, 429–435 (2008)
Dimas, A. S. et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science 325, 1246–1250 (2009)
Brem, R. B., Yvert, G., Clinton, R. & Kruglyak, L. Genetic dissection of transcriptional regulation in budding yeast. Science 296, 752–755 (2002)
Yvert, G. et al. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nature Genet. 35, 57–64 (2003)
Roider, H. G., Manke, T., O’Keeffe, S., Vingron, M. & Haas, S. A. PASTAA: identifying transcription factors associated with sets of co-regulated genes. Bioinformatics 25, 435–442 (2009)
Petretto, E. et al. New insights into the genetic control of gene expression using a Bayesian multi-tissue approach. PLoS Comput. Biol. 6, e1000737 (2010)
Breitling, R. et al. Genetical genomics: spotlight on QTL hotspots. PLoS Genet. 4, e1000232 (2008)
Nathan, C. & Ding, A. Nonresolving inflammation. Cell 140, 871–882 (2010)
Eizirik, D. L., Colli, M. L. & Ortis, F. The role of inflammation in insulitis and β-cell loss in type 1 diabetes. Nature Rev. Endocrinol. 5, 219–226 (2009)
Holness, C. L. & Simmons, D. L. Molecular cloning of CD68, a human macrophage marker related to lysosomal glycoproteins. Blood 81, 1607–1613 (1993)
Saar, K. et al. SNP and haplotype mapping for genetic analysis in the rat. Nature Genet. 40, 560–566 (2008)
Atanur, S. S. et al. The genome sequence of the spontaneously hypertensive rat: analysis and functional significance. Genome Res. 20, 791–803 (2010)
Zeller, T. et al. Genetics and beyond–the transcriptome of human monocytes and disease susceptibility. PLoS ONE 5, e10693 (2010)
Plagnol, V., Smyth, D. J., Todd, J. A. & Clayton, D. G. Statistical independence of the colocalized association signals for type 1 diabetes and RPS26 gene expression on chromosome 12q13. Biostatistics 10, 327–334 (2009)
von Herrath, M. Diabetes: a virus–gene collaboration. Nature 459, 518–519 (2009)
Nejentsev, S., Walker, N., Riches, D., Egholm, M. & Todd, J. A. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science 324, 387–389 (2009)
Smyth, D. J. et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nature Genet. 38, 617–619 (2006)
Barrett, J. C. et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nature Genet. 41, 703–707 (2009)
Li, Q. et al. Interferon-α initiates type 1 diabetes in nonobese diabetic mice. Proc. Natl Acad. Sci. USA 105, 12439–12444 (2008)
Cooper, J. D. et al. Follow-up of 1715 SNPs from the Wellcome Trust Case Control Consortium genome-wide association study in type I diabetes families. Genes Immun. 10 (suppl. 1). S85–S94 (2009)
Kawai, T. et al. Interferon-α induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nature Immunol. 5, 1061–1068 (2004)
The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007)
Acknowledgements
We acknowledge funding from the German National Genome Research Network (NGFN-Plus ‘Genetics of Heart Failure’), the Helmholtz Association Alliance on Systems Biology (MSBN), EURATools (LSHG-CT-2005-019015), European Union FP6 (LSHM-CT-2006-037593), PHC ALLIANCE 2009 (19419PH), UK National Institute for Health Research Biomedical Research Unit (Royal Brompton and Harefield NHS Trusts, University Hospitals of Leicester NHS Trusts) and Biomedical Research Centre (Imperial College NHS Trust) awards, the British Heart Foundation, grant P301/10/0290 from the Grant Agency of the Czech Republic, grant 1M6837805002 from the Ministry of Education of the Czech Republic, the Fondation Leducq, the Medical Research Council UK, Research Councils UK, the Juvenile Diabetes Research Foundation International, National Institute for Health Research (UK), National Institute of Diabetes and Digestive and Kidney Diseases (USA), and the Wellcome Trust. The research leading to these results has received funding from the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement no. HEALTH-F4-2010-241504 (EURATRANS). O. Burren performed T1DBase analyses.
Author information
Authors and Affiliations
Consortia
Contributions
S.A.C., N.H. and E.P. initiated the study. M.H., E.P., N.H. and S.A.C. participated in the conception, design and coordination of the study. H.L., Y.L., R.S., Y.A.L., S.P., C.R., K.S. and R.B. performed genetic, biochemical and functional analyses in rats. E.E.G. and J.G.C. provided Ebi2GFP/+ mouse data. M.P. and T.J.A. contributed materials and discussion of the manuscript. M.H., E.P., C.W., D.J.S., D.C., A.B., S.R.L., L.B., M.R. and L.T. designed and applied the modelling methodology and statistical analyses. M.H., E.P. and H. Schulz performed eQTL analysis in the rat. L.B. designed and performed the Bayesian analysis. C.W., D.J.S. and D.C. performed association analyses in humans. M.H., O.H., H.R. and M.V. designed and performed bioinformatics analyses in rats. J.E., C.H., S.M., W.H.O., C.M.R., N.J.S., H. Schunkert, A.H.G., S.B., T.M., T.Z., S.S., A.Z., M.R., L.T. and F.C. provided the human monocyte expression data and contributed to the transcriptomic analyses in the Cardiogenics Study and Gutenberg Heart Study cohorts. M.H., E.P., N.H. and S.A.C. wrote the paper with significant contributions from C.W. and J.A.T. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Microarray expression data in the rat have been deposited at ArrayExpress with the following identity codes: skeletal muscle, E-TABM-458; aorta, E-MTAB-322; liver, E-MTAB-323.
A list of participants and their affiliations appears at the end of the paper.
Supplementary information
Supplementary Information
This file contains Supplementary Information and Data, additional references and a list of contributors to the Cardiogenics Transcriptomic Study. (PDF 403 kb)
Supplementary Figures
This file contains Supplementary Figures 1-9 with legends. (PDF 2461 kb)
Supplementary Tables
This file contains Supplementary Tables 1-10. (PDF 1077 kb)
Rights and permissions
About this article
Cite this article
Heinig, M., Petretto, E., Wallace, C. et al. A trans-acting locus regulates an anti-viral expression network and type 1 diabetes risk. Nature 467, 460–464 (2010). https://doi.org/10.1038/nature09386
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09386
This article is cited by
-
A trans locus causes a ribosomopathy in hypertrophic hearts that affects mRNA translation in a protein length-dependent fashion
Genome Biology (2021)
-
Identification of biomarkers associated with metabolic cardiovascular disease using mRNA-SNP-miRNA regulatory network analysis
BMC Cardiovascular Disorders (2021)
-
CpGmotifs: a tool to discover DNA motifs associated to CpG methylation events
BMC Bioinformatics (2021)
-
Diabetes and Cardiovascular Complications: The Epidemics Continue
Current Cardiology Reports (2021)
-
The Roles of Orphan G Protein-Coupled Receptors in Autoimmune Diseases
Clinical Reviews in Allergy & Immunology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.