Abstract
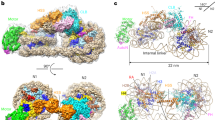
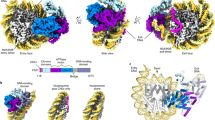
The small GTPase Ran enzyme regulates critical eukaryotic cellular functions including nuclear transport and mitosis through the creation of a RanGTP gradient around the chromosomes. This concentration gradient is created by the chromatin-bound RCC1 (regulator of chromosome condensation) protein, which recruits Ran to nucleosomes and activates Ran’s nucleotide exchange activity. Although RCC1 has been shown to bind directly with the nucleosome, the molecular details of this interaction were not known. Here we determine the crystal structure of a complex of Drosophila RCC1 and the nucleosome core particle at 2.9 Å resolution, providing an atomic view of how a chromatin protein interacts with the histone and DNA components of the nucleosome. Our structure also suggests that the Widom 601 DNA positioning sequence present in the nucleosomes forms a 145-base-pair nucleosome core particle, not the expected canonical 147-base-pair particle.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Carazo-Salas, R. E. et al. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature 400, 178–181 (1999)
Clarke, P. R. & Zhang, C. Spatial and temporal coordination of mitosis by Ran GTPase. Nature Rev. Mol. Cell Biol. 9, 464–477 (2008)
Kalab, P. & Heald, R. The RanGTP gradient—a GPS for the mitotic spindle. J. Cell Sci. 121, 1577–1586 (2008)
Renault, L. et al. The 1.7 Å crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature 392, 97–101 (1998)
Nemergut, M. E., Mizzen, C. A., Stukenberg, T., Allis, C. D. & Macara, I. G. Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science 292, 1540–1543 (2001)
Chen, T. et al. N-terminal α-methylation of RCC1 is necessary for stable chromatin association and normal mitosis. Nature Cell Biol. 9, 596–603 (2007)
Hao, Y. & Macara, I. G. Regulation of chromatin binding by a conformational switch in the tail of the Ran exchange factor RCC1. J. Cell Biol. 182, 827–836 (2008)
Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 (1997)
Bao, Y., White, C. L. & Luger, K. Nucleosome core particles containing a poly(dA.dT) sequence element exhibit a locally distorted DNA structure. J. Mol. Biol. 361, 617–624 (2006)
Chakravarthy, S. & Luger, K. The histone variant macro-H2A preferentially forms ‘hybrid nucleosomes’. J. Biol. Chem. 281, 25522–25531 (2006)
Clapier, C. R. et al. Structure of the Drosophila nucleosome core particle highlights evolutionary constraints on the H2A–H2B histone dimer. Proteins 71, 1–7 (2008)
Davey, C. A., Sargent, D. F., Luger, K., Maeder, A. W. & Richmond, T. J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 319, 1097–1113 (2002)
Ong, M. S., Richmond, T. J. & Davey, C. A. DNA stretching and extreme kinking in the nucleosome core. J. Mol. Biol. 368, 1067–1074 (2007)
Richmond, T. J. & Davey, C. A. The structure of DNA in the nucleosome core. Nature 423, 145–150 (2003)
Suto, R. K., Clarkson, M. J., Tremethick, D. J. & Luger, K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nature Struct. Biol. 7, 1121–1124 (2000)
Tsunaka, Y., Kajimura, N., Tate, S. & Morikawa, K. Alteration of the nucleosomal DNA path in the crystal structure of a human nucleosome core particle. Nucleic Acids Res. 33, 3424–3434 (2005)
White, C. L., Suto, R. K. & Luger, K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 20, 5207–5218 (2001)
Barbera, A. J. et al. The nucleosomal surface as a docking station for Kaposi’s sarcoma herpesvirus LANA. Science 311, 856–861 (2006)
Suto, R. K. et al. Crystal structures of nucleosome core particles in complex with minor groove DNA-binding ligands. J. Mol. Biol. 326, 371–380 (2003)
Lowary, P. T. & Widom, J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 (1998)
Davis, T. L., Bonacci, T. M., Sprang, S. R. & Smrcka, A. V. Structural and molecular characterization of a preferred protein interaction surface on G protein βγ subunits. Biochemistry 44, 10593–10604 (2005)
Lodowski, D. T., Pitcher, J. A., Capel, W. D., Lefkowitz, R. J. & Tesmer, J. J. Keeping G proteins at bay: a complex between G protein-coupled receptor kinase 2 and Gβγ. Science 300, 1256–1262 (2003)
Orlicky, S., Tang, X., Willems, A., Tyers, M. & Sicheri, F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112, 243–256 (2003)
Renault, L., Kuhlmann, J., Henkel, A. & Wittinghofer, A. Structural basis for guanine nucleotide exchange on Ran by the regulator of chromosome condensation (RCC1). Cell 105, 245–255 (2001)
England, J. R., Huang, J., Jennings, M. J., Makde, R. D. & Tan, S. RCC1 uses a conformationally diverse loop region to interact with the nucleosome: a model for the RCC1–nucleosome complex. J. Mol. Biol. 398, 518–529 (2010)
Dorigo, B. et al. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science 306, 1571–1573 (2004)
Shogren-Knaak, M. et al. Histone H4–K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847 (2006)
Koerber, R. T., Rhee, H. S., Jiang, C. & Pugh, B. F. Interaction of transcriptional regulators with specific nucleosomes across the Saccharomyces genome. Mol. Cell 35, 889–902 (2009)
Edayathumangalam, R. S., Weyermann, P., Dervan, P. B., Gottesfeld, J. M. & Luger, K. Nucleosomes in solution exist as a mixture of twist-defect states. J. Mol. Biol. 345, 103–114 (2005)
Gangaraju, V. K., Prasad, P., Srour, A., Kagalwala, M. N. & Bartholomew, B. Conformational changes associated with template commitment in ATP-dependent chromatin remodeling by ISW2. Mol. Cell 35, 58–69 (2009)
Saha, A., Wittmeyer, J. & Cairns, B. R. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nature Struct. Mol. Biol. 12, 747–755 (2005)
Bilbao-Cortes, D., Hetzer, M., Langst, G., Becker, P. B. & Mattaj, I. W. Ran binds to chromatin by two distinct mechanisms. Curr. Biol. 12, 1151–1156 (2002)
Zhang, C., Goldberg, M. W., Moore, W. J., Allen, T. D. & Clarke, P. R. Concentration of Ran on chromatin induces decondensation, nuclear envelope formation and nuclear pore complex assembly. Eur. J. Cell Biol. 81, 623–633 (2002)
Partridge, J. R. & Schwartz, T. U. Crystallographic and biochemical analysis of the Ran-binding zinc finger domain. J. Mol. Biol. 391, 375–389 (2009)
Chook, Y. M. & Blobel, G. Structure of the nuclear transport complex karyopherin-β2–Ran·GppNHp. Nature 399, 230–237 (1999)
Seewald, M. J., Korner, C., Wittinghofer, A. & Vetter, I. R. RanGAP mediates GTP hydrolysis without an arginine finger. Nature 415, 662–666 (2002)
Vetter, I. R., Nowak, C., Nishimoto, T., Kuhlmann, J. & Wittinghofer, A. Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature 398, 39–46 (1999)
Otwinoski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004)
Delano, W. L. The PyMOL molecular graphics system 〈http://www.pymol.org〉.
Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA 98, 10037–10041 (2001)
Luger, K., Rechsteiner, T. J. & Richmond, T. J. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 119, 1–16 (1999)
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK—a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283–291 (1993)
Brunger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998)
Lavery, R., Moakher, M., Maddocks, J. H., Petkeviciute, D. & Zakrzewska, K. Conformational analysis of nucleic acids revisited: Curves+. Nucleic Acids Res. 37, 5917–5929 (2009)
Kleywegt, G. J. Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr. D Biol. Crystallogr. 52, 842–857 (1996)
Tan, S., Kern, R. C. & Selleck, W. The pST44 polycistronic expression system for producing protein complexes in Escherichia coli . Protein Expr. Purif. 40, 385–395 (2005)
Acknowledgements
We thank M. Saxena for sharing unpublished coordinates of Drosophila RCC1; K. Wiley, D. Schlaich and M. Porzio for technical assistance; the staff of APS NE-CAT beamline 24-ID-E and Cornell CHESS beamlines A1 and F1 for their assistance during synchrotron data collection; N. Yennawar at the Penn Sate Huck Institutes X-ray core facility; W. Selleck, M. Adams, the members of the Tan laboratory and the Penn State Center for Eukaryotic Gene Regulation for discussions; T. Stukenberg for advice and encouragement at the initiation of this project, J. Widom for sending the 601 nucleosome DNA positioning sequence; and the Pew Scholar 20th Reunion Meeting for stimulating this project.
Author information
Authors and Affiliations
Contributions
R.D.M. cloned and purified macromolecules, crystallized, collected, processed X-ray data, refined and analysed the structure. J.R.E. performed pulldown assays and collected X-ray data. H.P.Y. collected and processed X-ray data. S.T. designed the study, cloned and purified macromolecules, crystallized, collected X-ray data, analysed the results and wrote the paper. All authors commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-5 with legends, Supplementary Table 1 and Supplementary Results. (PDF 948 kb)
Rights and permissions
About this article
Cite this article
Makde, R., England, J., Yennawar, H. et al. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature 467, 562–566 (2010). https://doi.org/10.1038/nature09321
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09321
This article is cited by
-
Lola-I is a promoter pioneer factor that establishes de novo Pol II pausing during development
Nature Communications (2023)
-
RCCD1 promotes breast carcinogenesis through regulating hypoxia-associated mitochondrial homeostasis
Oncogene (2023)
-
CENP-A and CENP-B collaborate to create an open centromeric chromatin state
Nature Communications (2023)
-
Structural basis of paralog-specific KDM2A/B nucleosome recognition
Nature Chemical Biology (2023)
-
Tandem mass tag (TMT)-based proteomic analysis of Cryptosporidium andersoni oocysts before and after excystation
Parasites & Vectors (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.