Abstract
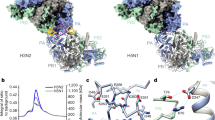
The heterotrimeric influenza virus polymerase, containing the PA, PB1 and PB2 proteins, catalyses viral RNA replication and transcription in the nucleus of infected cells. PB1 holds the polymerase active site1 and reportedly harbours endonuclease activity2, whereas PB2 is responsible for cap binding2,3,4. The PA amino terminus is understood to be the major functional part of the PA protein and has been implicated in several roles, including endonuclease5 and protease activities6 as well as viral RNA/complementary RNA promoter binding7. Here we report the 2.2 ångström (Å) crystal structure of the N-terminal 197 residues of PA, termed PAN, from an avian influenza H5N1 virus. The PAN structure has an α/β architecture and reveals a bound magnesium ion coordinated by a motif similar to the (P)DXN(D/E)XK motif characteristic of many endonucleases. Structural comparisons and mutagenesis analysis of the motif identified in PAN provide further evidence that PAN holds an endonuclease active site. Furthermore, functional analysis with in vivo ribonucleoprotein reconstitution and direct in vitro endonuclease assays strongly suggest that PAN holds the endonuclease active site and has critical roles in endonuclease activity of the influenza virus polymerase, rather than PB1. The high conservation of this endonuclease active site among influenza strains indicates that PAN is an important target for the design of new anti-influenza therapeutics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Poch, O., Sauvaget, I., Delarue, M. & Tordo, N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8, 3867–3874 (1989)
Li, M. L., Rao, P. & Krug, R. M. The active sites of the influenza cap-dependent endonuclease are on different polymerase subunits. EMBO J. 20, 2078–2086 (2001)
Guilligay, D. et al. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nature Struct. Mol. Biol. 15, 500–506 (2008)
Fechter, P. et al. Two aromatic residues in the PB2 subunit of influenza A RNA polymerase are crucial for cap binding. J. Biol. Chem. 278, 20381–20388 (2003)
Hara, K., Schmidt, F. I., Crow, M. & Brownlee, G. G. Amino acid residues in the N-terminal region of the PA subunit of influenza A virus RNA polymerase play a critical role in protein stability, endonuclease activity, cap binding, and virion RNA promoter binding. J. Virol. 80, 7789–7798 (2006)
Sanz-Ezquerro, J. J., Zurcher, T., de la Luna, S., Ortin, J. & Nieto, A. The amino-terminal one-third of the influenza virus PA protein is responsible for the induction of proteolysis. J. Virol. 70, 1905–1911 (1996)
Maier, H. J., Kashiwagi, T., Hara, K. & Brownlee, G. G. Differential role of the influenza A virus polymerase PA subunit for vRNA and cRNA promoter binding. Virology 370, 194–204 (2008)
Lee, M. T. et al. Definition of the minimal viral components required for the initiation of unprimed RNA synthesis by influenza virus RNA polymerase. Nucleic Acids Res. 30, 429–438 (2002)
Regan, J. F., Liang, Y. & Parslow, T. G. Defective assembly of influenza A virus due to a mutation in the polymerase subunit PA. J. Virol. 80, 252–261 (2006)
Naffakh, N., Massin, P. & van der Werf, S. The transcription/replication activity of the polymerase of influenza A viruses is not correlated with the level of proteolysis induced by the PA subunit. Virology 285, 244–252 (2001)
Fodor, E. et al. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 76, 8989–9001 (2002)
Hara, K. et al. Influenza virus RNA polymerase PA subunit is a novel serine protease with Ser624 at the active site. Genes Cells 6, 87–97 (2001)
Zurcher, T., de la Luna, S., Sanz-Ezquerro, J. J., Nieto, A. & Ortin, J. Mutational analysis of the influenza virus A/Victoria/3/75 PA protein: studies of interaction with PB1 protein and identification of a dominant negative mutant. J. Gen. Virol. 77, 1745–1749 (1996)
Huarte, M. et al. Threonine 157 of influenza virus PA polymerase subunit modulates RNA replication in infectious viruses. J. Virol. 77, 6007–6013 (2003)
Kawaguchi, A., Naito, T. & Nagata, K. Involvement of influenza virus PA subunit in assembly of functional RNA polymerase complexes. J. Virol. 79, 732–744 (2005)
Guu, T. S., Dong, L., Wittung-Stafshede, P. & Tao, Y. J. Mapping the domain structure of the influenza A virus polymerase acidic protein (PA) and its interaction with the basic protein 1 (PB1) subunit. Virology 379, 135–142 (2008)
He, X. et al. Crystal structure of the polymerase PA(C)-PB1(N) complex from an avian influenza H5N1 virus. Nature 454, 1123–1126 (2008)
Obayashi, E. et al. The structural basis for an essential subunit interaction in influenza virus RNA polymerase. Nature 454, 1127–1131 (2008)
Blok, V. et al. Inhibition of the influenza virus RNA-dependent RNA polymerase by antisera directed against the carboxy-terminal region of the PB2 subunit. J. Gen. Virol. 77, 1025–1033 (1996)
Honda, A., Mizumoto, K. & Ishihama, A. Minimum molecular architectures for transcription and replication of the influenza virus. Proc. Natl Acad. Sci. USA 99, 13166–13171 (2002)
Shi, L., Summers, D. F., Peng, Q. & Galarz, J. M. Influenza A virus RNA polymerase subunit PB2 is the endonuclease which cleaves host cell mRNA and functions only as the trimeric enzyme. Virology 208, 38–47 (1995)
Newman, M., Strzelecka, T., Dorner, L. F., Schildkraut, I. & Aggarwal, A. K. Structure of restriction endonuclease BamHI and its relationship to EcoRI. Nature 368, 660–664 (1994)
Lukacs, C. M., Kucera, R., Schildkraut, I. & Aggarwal, A. K. Understanding the immutability of restriction enzymes: crystal structure of BglII and its DNA substrate at 1.5 A resolution. Nature Struct. Biol. 7, 134–140 (2000)
Fodor, E. & Smith, M. The PA subunit is required for efficient nuclear accumulation of the PB1 subunit of the influenza A virus RNA polymerase complex. J. Virol. 78, 9144–9153 (2004)
Nieto, A. et al. Nuclear transport of influenza virus polymerase PA protein. Virus Res. 24, 65–75 (1992)
Nieto, A., de la Luna, S., Barcena, J., Portela, A. & Ortin, J. Complex structure of the nuclear translocation signal of influenza virus polymerase PA subunit. J. Gen. Virol. 75, 29–36 (1994)
Perales, B. et al. The replication activity of influenza virus polymerase is linked to the capacity of the PA subunit to induce proteolysis. J. Virol. 74, 1307–1312 (2000)
Toyoda, T., Adyshev, D. M., Kobayashi, M., Iwata, A. & Ishihama, A. Molecular assembly of the influenza virus RNA polymerase: determination of the subunit-subunit contact sites. J. Gen. Virol. 77, 2149–2157 (1996)
Vreede, F. T., Jung, T. E. & Brownlee, G. G. Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates. J. Virol. 78, 9568–9572 (2004)
Brownlee, G. G., Fodor, E., Pritlove, D. C., Gould, K. G. & Dalluge, J. J. Solid phase synthesis of 5′-diphosphorylated oligoribonucleotides and their conversion to capped m7Gppp-oligoribonucleotides for use as primers for influenza A virus RNA polymerase in vitro . Nucleic Acids Res. 23, 2641–2647 (1995)
Otwinowski, Z. & Minor, W. in Macromolecular Crystallography Vol. 276 (eds Carter, C. W. Jr & Sweet, R. M.) Part A 307–326 (Academic, 1997)
Sheldrick, G. M. A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008)
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Read, R. J. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D 57, 1373–1382 (2001)
Brunger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Acknowledgements
We thank H. Chen and K. Yu for providing the A/goose/Guangdong/1/96 influenza PA gene, R. Zhang and A. Joachimiak for assistance with data collection, and G. G. Brownlee for providing materials and for valuable advice. This work was supported by the National Natural Science Foundation of China (grant numbers 30599432, 30221003 and 30721003), the Ministry of Science and Technology International Cooperation Project (2006DFB32420), the Ministry of Science and Technology 863 Project (2006AA02A314 and 2006AA02A322), the Ministry of Science and Technology 973 Project (2006CB504300 and 2007CB914300) and the Medical Research Council (G0700848).
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Information
This file contains Supplementary Figures S1-S4 with Legends and Supplementary Table T1. (PDF 449 kb)
Rights and permissions
About this article
Cite this article
Yuan, P., Bartlam, M., Lou, Z. et al. Crystal structure of an avian influenza polymerase PAN reveals an endonuclease active site. Nature 458, 909–913 (2009). https://doi.org/10.1038/nature07720
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07720
This article is cited by
-
Mechanisms and consequences of mRNA destabilization during viral infections
Virology Journal (2024)
-
Exploring the alternative virulence determinants PB2 S155N and PA S49Y/D347G that promote mammalian adaptation of the H9N2 avian influenza virus in mice
Veterinary Research (2023)
-
Cut site preference allows influenza A virus PA-X to discriminate between host and viral mRNAs
Nature Microbiology (2023)
-
The ubiquitination landscape of the influenza A virus polymerase
Nature Communications (2023)
-
Host adaptive mutations in the 2009 H1N1 pandemic influenza A virus PA gene regulate translation efficiency of viral mRNAs via GRSF1
Communications Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.