Abstract
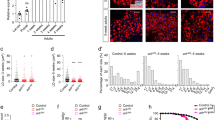
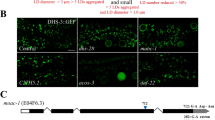
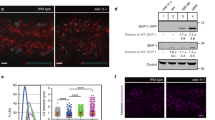
Eukaryotic cells store neutral lipids in cytoplasmic lipid droplets1,2 enclosed in a monolayer of phospholipids and associated proteins3,4. These dynamic organelles5 serve as the principal reservoirs for storing cellular energy and for the building blocks for membrane lipids. Excessive lipid accumulation in cells is a central feature of obesity, diabetes and atherosclerosis, yet remarkably little is known about lipid-droplet cell biology. Here we show, by means of a genome-wide RNA interference (RNAi) screen in Drosophila S2 cells that about 1.5% of all genes function in lipid-droplet formation and regulation. The phenotypes of the gene knockdowns sorted into five distinct phenotypic classes. Genes encoding enzymes of phospholipid biosynthesis proved to be determinants of lipid-droplet size and number, suggesting that the phospholipid composition of the monolayer profoundly affects droplet morphology and lipid utilization. A subset of the Arf1–COPI vesicular transport proteins also regulated droplet morphology and lipid utilization, thereby identifying a previously unrecognized function for this machinery. These phenotypes are conserved in mammalian cells, suggesting that insights from these studies are likely to be central to our understanding of human diseases involving excessive lipid storage.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bartz, R. et al. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J. Lipid Res. 48, 837–847 (2007)
Brown, D. A. Lipid droplets: proteins floating on a pool of fat. Curr. Biol. 11, R446–R449 (2001)
Miura, S. et al. Functional conservation for lipid storage droplet association among Perilipin, ADRP, and TIP47 (PAT)-related proteins in mammals, Drosophila, and Dictyostelium. J. Biol. Chem. 277, 32253–32257 (2002)
Bartz, R. et al. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J. Proteome Res. 6, 3256–3265 (2007)
Martin, S. & Parton, R. G. Lipid droplets: a unified view of a dynamic organelle. Nature Rev. Mol. Cell Biol. 7, 373–378 (2006)
Ulvila, J. et al. Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in Drosophila S2 cells. J. Biol. Chem. 281, 14370–14375 (2006)
Shaner, N. C. et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature Biotechnol. 22, 1567–1572 (2004)
Ma, Y., Creanga, A., Lum, L. & Beachy, P. A. Prevalence of off-target effects in Drosophila RNA interference screens. Nature 443, 359–363 (2006)
Kulkarni, M. M. et al. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nature Methods 3, 833–838 (2006)
Goshima, G. et al. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316, 417–421 (2007)
Brown, M. S. & Goldstein, J. L. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc. Natl Acad. Sci. USA 96, 11041–11048 (1999)
Dobrosotskaya, I. Y., Seegmiller, A. C., Brown, M. S., Goldstein, J. L. & Rawson, R. B. Regulation of SREBP processing and membrane lipid production by phospholipids in Drosophila. Science 296, 879–883 (2002)
Kent, C. Regulatory enzymes of phosphatidylcholine biosynthesis: a personal perspective. Biochim. Biophys. Acta 1733, 53–66 (2005)
Morrison, D. K., Murakami, M. S. & Cleghon, V. Protein kinases and phosphatases in the Drosophila genome. J. Cell Biol. 150, 57–62 (2000)
Cornell, R. B. & Northwood, I. C. Regulation of CTP:phosphocholine cytidylyltransferase by amphitropism and relocalization. Trends Biochem. Sci. 25, 441–447 (2000)
Weber, U., Eroglu, C. & Mlodzik, M. Phospholipid membrane composition affects EGF receptor and Notch signaling through effects on endocytosis during Drosophila development. Dev. Cell 5, 559–570 (2003)
Hafez, I. M. & Cullis, P. R. Roles of lipid polymorphism in intracellular delivery. Adv. Drug Deliv. Rev. 47, 139–148 (2001)
Dascher, C. & Balch, W. E. Dominant inhibitory mutants of ARF1 block endoplasmic reticulum to Golgi transport and trigger disassembly of the Golgi apparatus. J. Biol. Chem. 269, 1437–1448 (1994)
Nakamura, N., Banno, Y. & Tamiya-Koizumi, K. Arf1-dependent PLD1 is localized to oleic acid-induced lipid droplets in NIH3T3 cells. Biochem. Biophys. Res. Commun. 335, 117–123 (2005)
Spang, A., Matsuoka, K., Hamamoto, S., Schekman, R. & Orci, L. Coatomer, Arf1p, and nucleotide are required to bud coat protein complex I-coated vesicles from large synthetic liposomes. Proc. Natl Acad. Sci. USA 95, 11199–11204 (1998)
D'Souza-Schorey, C. & Chavrier, P. ARF proteins: roles in membrane traffic and beyond. Nature Rev. Mol. Cell Biol. 7, 347–358 (2006)
Marcinkiewicz, A., Gauthier, D., Garcia, A. & Brasaemle, D. L. The phosphorylation of serine 492 of perilipin a directs lipid droplet fragmentation and dispersion. J. Biol. Chem. 281, 11901–11909 (2006)
Verghese, P. B., Arrese, E. L. & Soulages, J. L. Stimulation of lipolysis enhances the rate of cholesterol efflux to HDL in adipocytes. Mol. Cell. Biochem. 302, 241–248 (2007)
Bard, F. et al. Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature 439, 604–607 (2006)
Kumar, Y., Cocchiaro, J. & Valdivia, R. H. The obligate intracellular pathogen Chlamydia trachomatis targets host lipid droplets. Curr. Biol. 16, 1646–1651 (2006)
Miyanari, Y. et al. The lipid droplet is an important organelle for hepatitis C virus production. Nature Cell Biol. 9, 1089–1097 (2007)
Goshima, G. & Vale, R. D. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 162, 1003–1016 (2003)
Rogers, S. L., Wiedemann, U., Stuurman, N. & Vale, R. D. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162, 1079–1088 (2003)
Hojjati, M. R. & Jiang, X. C. Rapid, specific, and sensitive measurements of plasma sphingomyelin and phosphatidylcholine. J. Lipid Res. 47, 673–676 (2006)
Monetti, M. et al. Dissociation of hepatic steatosis and insulin resistance in mice overexpressing DGAT in the liver. Cell Metab. 6, 69–78 (2007)
Acknowledgements
We thank I. Vasenkova and R. De Breuil for help with dsRNA synthesis; K. Warner for help with yeast work; M. Schuldiner, E. Griffis, T. Fazzio, E. Herker, S. Stymne, B. Panning and M. Ott for reagents; D. B. Jones and G. Howard for assistance with manuscript preparation; G. Schoenhofer for web access for the database; members of the Farese, Vale and Walter laboratories for discussions; and D. Srivastava and S. Yamanaka for critical reading of the manuscript. This work was supported by a Freedom to Discover Award from Bristol–Myers Squibb and National Institutes of Health grant R21 DK078254-01 (to R.F.), a David and Mary Phillips postdoctoral fellowship award (to Y.G.), the Human Frontier Science Program Organization (T.C.W.), the Howard Hughes Medical Institute (P.W. and R.D.V.) and the J. David Gladstone Institutes.
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary information
The file contains Supplementary Methods, Supplementary Tables S1-S3 and Supplementary Figures S1-S5. (PDF 4198 kb)
Supplementary information
The file contains Supplementary Movie 1 (AVI 3709 kb)
Supplementary information
The file contains Supplementary Movie 2. (AVI 27099 kb)
Rights and permissions
About this article
Cite this article
Guo, Y., Walther, T., Rao, M. et al. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature 453, 657–661 (2008). https://doi.org/10.1038/nature06928
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature06928
This article is cited by
-
Nontargeted metabolomics reveals the potential mechanism underlying the association between birthweight and metabolic disturbances
BMC Pregnancy and Childbirth (2023)
-
Lipid droplet biogenesis and functions in health and disease
Nature Reviews Endocrinology (2023)
-
CLSTN3β enforces adipocyte multilocularity to facilitate lipid utilization
Nature (2023)
-
Glutathione S-transferase Mu 2 inhibits hepatic steatosis via ASK1 suppression
Communications Biology (2022)
-
Circadian Regulation and Clock-Controlled Mechanisms of Glycerophospholipid Metabolism from Neuronal Cells and Tissues to Fibroblasts
Molecular Neurobiology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.