Abstract
Suicides are a leading cause of death in psychiatric patients, and in society at large. Developing more quantitative and objective ways (biomarkers) for predicting and tracking suicidal states would have immediate practical applications and positive societal implications. We undertook such an endeavor. First, building on our previous blood biomarker work in mood disorders and psychosis, we decided to identify blood gene expression biomarkers for suicidality, looking at differential expression of genes in the blood of subjects with a major mood disorder (bipolar disorder), a high-risk population prone to suicidality. We compared no suicidal ideation (SI) states and high SI states using a powerful intrasubject design, as well as an intersubject case–case design, to generate a list of differentially expressed genes. Second, we used a comprehensive Convergent Functional Genomics (CFG) approach to identify and prioritize from the list of differentially expressed gene biomarkers of relevance to suicidality. CFG integrates multiple independent lines of evidence—genetic and functional genomic data—as a Bayesian strategy for identifying and prioritizing findings, reducing the false-positives and false-negatives inherent in each individual approach. Third, we examined whether expression levels of the blood biomarkers identified by us in the live bipolar subject cohort are actually altered in the blood in an age-matched cohort of suicide completers collected from the coroner’s office, and report that 13 out of the 41 top CFG scoring biomarkers (32%) show step-wise significant change from no SI to high SI states, and then to the suicide completers group. Six out of them (15%) remained significant after strict Bonferroni correction for multiple comparisons. Fourth, we show that the blood levels of SAT1 (spermidine/spermine N1–acetyltransferase 1), the top biomarker identified by us, at the time of testing for this study, differentiated future as well as past hospitalizations with suicidality, in a live cohort of bipolar disorder subjects, and exhibited a similar but weaker pattern in a live cohort of psychosis (schizophrenia/schizoaffective disorder) subjects. Three other (phosphatase and tensin homolog (PTEN), myristoylated alanine-rich protein kinase C substrate (MARCKS), and mitogen-activated protein kinase kinase kinase 3 (MAP3K3)) of the six biomarkers that survived Bonferroni correction showed similar but weaker effects. Taken together, the prospective and retrospective hospitalization data suggests SAT1, PTEN, MARCKS and MAP3K3 might be not only state biomarkers but trait biomarkers as well. Fifth, we show how a multi-dimensional approach using SAT1 blood expression levels and two simple visual-analog scales for anxiety and mood enhances predictions of future hospitalizations for suicidality in the bipolar cohort (receiver-operating characteristic curve with area under the curve of 0.813). Of note, this simple approach does not directly ask about SI, which some individuals may deny or choose not to share with clinicians. Lastly, we conducted bioinformatic analyses to identify biological pathways, mechanisms and medication targets. Overall, suicidality may be underlined, at least in part, by biological mechanisms related to stress, inflammation and apoptosis.
Similar content being viewed by others
Introduction
‘To be, or not to be, that is the question’W Shakespeare, Hamlet
Whatever its evolutionary, teleological and cultural reasons for existing, suicidal behavior is in most cases pathological and leads to irreversible tragedies.1, 2 Paradoxically, given its importance, there are yet no reliable objective tools to assess and track changes in suicidal risk without asking the individuals directly. Such tools are desperately needed, as individuals at risk often choose not to share their ideation or intent with others, for fear of stigma, hospitalization, or that in fact their plans may be thwarted.
A convergence of methods assessing the persons’ internal subjective feelings and thoughts, along with external, more objective ratings of actions and behaviors, are used de facto in clinical psychiatry. Such an approach is insufficient and is lagging behind those used in other medical specialties. It lacks precision, objectivity and predictive ability.
Our group has previously provided the first proof-of-principle for the use of blood gene expression biomarkers to predict mood state3 and psychosis symptoms.4 As the target organ in psychiatry—the brain—cannot be biopsied in live patients, it is essential to be able to identify and validate peripheral biomarkers for subsequent practical implementation in clinical settings. We now present a comprehensive and highly reductionist approach for discovering and validating blood biomarkers for suicidality.
We used a Convergent Functional Genomics (CFG) approach to identify and prioritize biomarkers of relevance to suicidality. CFG is a powerful, combined approach for extracting signal from noise in genetic and gene expression studies. The CFG methodology has already been applied to help identify and prioritize candidate genes, pathways and mechanisms for neuropsychiatric disorders, such as bipolar disorder,5, 6, 7, 8 alcoholism,9 anxiety10 and schizophrenia,11 showing reproducibility and predictive ability in independent cohorts.
Subjects and methods
Human subjects
We present data from four cohorts: one live bipolar discovery cohort; one postmortem coroner’s office test cohort; and two prospective follow-up live cohorts–one bipolar and one psychosis (schizophrenia/schizoaffective).
These live subjects are part of a larger longitudinal cohort being collected and studied by us. Subjects are recruited from the patient population at the Indianapolis VA Medical Center, the Indiana University School of Medicine, as well as various facilities that serve people with mental illnesses in Indiana. The subjects are recruited largely through referrals from care providers, the use of brochures left in plain sight in public places and mental health clinics, and through word of mouth. Subjects were excluded if they had significant medical or neurological illness or had evidence of active substance abuse or dependence. All subjects understood and signed informed consent forms detailing the research goals, procedure, caveats and safeguards. Subjects completed diagnostic assessments by an extensive structured clinical interview—Diagnostic Interview for Genetic Studies—at a baseline visit, followed by up to three testing visits, 3–6 months apart. At each testing visit, they received a series of psychiatric rating scales, including the Hamilton Rating Scale for Depression-17, which includes a suicidal ideation (SI) rating item (Figure 1), and the blood was drawn. Whole blood (10 ml) was collected in two RNA-stabilizing PAXgene tubes, labeled with an anonymized ID number, and stored at −80 °C in a locked freezer until the time of future processing. Whole-blood (predominantly lymphocyte) RNA was extracted for microarray gene expression studies from the PAXgene tubes, as detailed below. We focused this initial study on a male population because of the demographics of our catchment area (primarily male in a VA Medical Center), and to minimize any potential gender-related effects on gene expression, which would have decreased the discriminative power of our analysis given our relatively small sample size.
Discovery cohort: intrasubject and intersubject analyses. Phchp### is study ID for each subject. V# after it denotes visit number (1, 2 or 3). (a) Design and (b) suicidal ideation (SI) scoring. (c) Overlapping probesets and genes.
Our intrasubject discovery cohort, from which the biomarker data were derived, consisted of nine male Caucasian subjects with bipolar disorder, with multiple visits, who each had a diametric change in SI scores from no SI to high SI from one testing visit to another testing visit. There were 6 subjects with 3 visits each, and 3 subjects with 2 visits each, resulting in a total of 24 blood samples for subsequent microarray studies (Table 1 and Figure 1).
Our postmortem cohort, in which the top biomarker findings were tested, consisted of an age-matched cohort of nine male suicide completers obtained through the Marion County coroner’s office (eight Caucasians, one African American) (Table 1 and Supplementary Table S2). We required a last observed alive postmortem interval of 24 h or less, and the cases selected had completed suicide by means other than overdose, which could affect gene expression. Next of kin signed informed consent at the coroner’s office for donation of tissues and fluids for research. The samples were collected as part of our INBRAIN initiative (Indiana Center for Biomarker Research in Neuropsychiatry).
The bipolar follow-up cohort (n=42) (Table 1) consisted of male Caucasian subjects in whom whole-genome blood gene expression data, including levels of SAT1 (spermidine/spermine N1–acetyltransferase 1), were obtained by us at testing visits over the years as part of our longitudinal study. If the subjects had multiple testing visits, the visit with the highest SAT1 level was selected for this analysis. The subjects’ subsequent number of hospitalizations with or without suicidality was tabulated from electronic medical records. The psychosis (schizophrenia/schizoaffective) follow-up cohort (n=46) (Supplementary Table S9) similarly consisted of Caucasian subjects in whom whole-genome blood gene expression data, including levels of SAT1, were obtained by us at testing visits over the years as part of our longitudinal study. If the subjects had multiple testing visits, the visit with the highest SAT1 level was selected for this analysis. The subjects’ subsequent number of hospitalizations with or without suicidality was tabulated from electronic medical records. A hospitalization was deemed to be without suicidality if suicidality was not listed as a reason for admission, and no SI was described in the admission and discharge medical notes. Conversely, a hospitalization was deemed to be because of suicidality if suicidal acts or intent was listed as a reason for admission, and SI was described in the admission and discharge medical notes.
Medications
The subjects in the discovery cohort were all diagnosed with bipolar disorder (Table 1). Their psychiatric medications are listed in Supplementary Table S1. The subjects were on a variety of different psychiatric medications: mood stabilizer, antidepressants, antipsychotics, benzodiazepines and others. Medications can have a strong influence on gene expression. However, our discovery of differentially expressed genes was based on intrasubject analyses, which factor out not only genetic background effects but also medication effects, as the subjects had no major medication changes between visits. Moreover, there was no consistent pattern in any particular type of medication, or between any change in medications and SI, in the rare instances where there were changes in medications between visits.
Human blood gene expression experiments and analyses
RNA extraction
Whole blood (2.5–5 ml) was collected into each PaxGene tube by routine venipuncture. PaxGene tubes contain proprietary reagents for the stabilization of RNA. The cells from whole blood were concentrated by centrifugation, the pellet washed, resuspended and incubated in buffers containing Proteinase K for protein digestion. A second centrifugation step was done to remove residual cell debris. After the addition of ethanol for an optimal binding condition, the lysate was applied to a silica-gel membrane/column. The RNA bound to the membrane as the column was centrifuged, and contaminants were removed in three wash steps. The RNA was then eluted using diethylpyrocarbonate-treated water. The protocol for RNA extraction is carried out on a QIAgen QIAcube.
Sample labeling
Sample labeling was performed using the Ambion MessageAmp II-BiotinEnhanced antisense RNA (aRNA) amplification kit. The procedure is briefly outlined below and involves the following steps:
-
1
Reverse transcription to synthesize first-strand cDNA was primed with the T7 oligo(dT) primer to synthesize cDNA containing a T7 promoter sequence.
-
2
Second-strand cDNA synthesis converted the single-stranded cDNA into a double-stranded DNA template for transcription. The reaction employed DNA polymerase and RNase H to simultaneously degrade the RNA and synthesize the second-strand cDNA.
-
3
cDNA purification removed RNA, primers, enzymes and salts that would have inhibited in vitro transcription.
-
4
In vitro transcription to synthesize aRNA with biotin–NTP Mix generated multiple copies of biotin-modified aRNA from the double-stranded cDNA templates; this is the amplification step.
-
5
aRNA purification removed unincorporated NTPs, salts, enzymes and inorganic phosphate to improve the stability of the biotin-modified aRNA.
-
6
aRNA fragmentation: the amplified RNA is fragmented in a reaction that employs a metal-induced hydrolysis to fragment the aRNA. The fragmented labeled aRNA is now ready for hybridization to the Affymetrix microarray chip (Affymetrix, Santa Clara, CA, USA).
Microarrays
Biotin-labeled aRNAs were hybridized to Affymetrix HG-U133 Plus 2.0 GeneChips (Affymetrix; with over 40 000 genes and expressed sequence tags), according to the manufacturer’s protocols http://www.affymetrix.com/support/technical/manual/expression_manual.affx. Arrays were stained using standard Affymetrix protocols for antibody signal amplification and scanned on an Affymetrix GeneArray 2500 scanner with a target intensity set at 250. Quality-control measures, including 30/50 ratios for glyceraldehyde 3-phosphate dehydrogenase and β-actin, scale factors, background and Q–values, were within acceptable limits.
Analysis
We have used the subject’s SI scores at the time of blood collection (0—no SI compared with 2 and above—high SI). We looked at gene expression differences between the no SI and the high SI visits, using both an intrasubject and an intersubject design (Figure 1).
Differential gene expression analyses in the discovery cohort
We imported all Affymetrix microarray data as cel files into Partek Genomic Suites 6.6 software package (Partek Incorporated, St Louis, MI, USA). Using only the perfect match values, we ran a robust multi-array analysis (RMA), background corrected with quantile normalization and a median polish probeset summarization of all 24 chips, to obtain the normalized expression levels of all probesets for each chip. Then, to establish a list of differentially expressed probesets we ran two analyses.
An intrasubject analysis using a fold change in expression of at least 1.2 between high- and no SI visits within each subject was performed. There were in total 15 comparisons. Probesets that had a 1.2-fold change were then assigned either a 1 (increased in high SI) or a −1 (decreased in high SI) in each comparison. These values were then summed for each probeset across the 15 comparisons, yielding a range of scores between −11 and 12. The probesets in the top 5% (1269 probesets, <5% of 54 675 total probesets) had an absolute (without sign) score value of 7 and greater, and received an internal CFG score of 1 point. The probesets in the top 0.1% (24 probesets, <0.1% of 54 675 total probesets) had an absolute score of 11 and greater, and received an internal CFG score of 3 points.
In addition, an intersubject analysis using t-test (two-tailed, unequal variance) was performed to find probesets differentially expressed between high SI and no SI chips (Figure 1), resulting in 648 probesets with P<0.05. Probesets with a P<0.05 received an internal CFG score of 1 point, whereas probesets with P<0.001 received 3 points.
We further filtered results by only selecting probesets that overlapped between the intrasubject and the intersubject analyses, resulting in 279 probesets corresponding to 246 unique genes. Gene names for the probesets were identified using Partek and NetAffyx (Affymetrix) for Affymetrix HG-U133 Plus 2.0 GeneChips, followed by GeneCards to confirm the primary gene symbol. In addition, for those probesets that were not assigned a gene name by Partek or NetAffyx, we used the UCSC Genome Browser on Human Feb. 2009 (GRCh37/hg19) to directly map them to known genes. Genes were then scored using our manually curated CFG databases as described below (Figure 2).
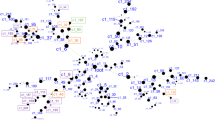
Convergent Functional Genomics approach for identification and prioritization of genomic biomarkers for suicidality.
Convergent Functional Genomics
Databases
We have established in our laboratory (Laboratory of Neurophenomics, Indiana University School of Medicine, www.neurophenomics.info) manually curated databases of all the human gene expression (postmortem brain, blood and cell cultures), human genetics (association, copy number variations and linkage), and animal model gene expression and genetic studies published to date on psychiatric disorders.12 Only the findings deemed significant in the primary publication, by the study authors, using their particular experimental design and thresholds, are included in our databases. Our databases include only primary literature data and do not include review papers or other secondary data integration analyses to avoid redundancy and circularity. These large and constantly updated databases have been used in our CFG cross validation and prioritization (Figure 2).
Human postmortem brain gene expression evidence
Information about genes was obtained and imported in our databases by searching the primary literature with PubMed (http://ncbi.nlm.nih.gov/PubMed), using various combinations of keywords (gene name, suicide, suicide gene expression and human brain). Postmortem convergence was deemed to occur for a gene if there were published reports of human postmortem data showing changes in expression of that gene in brains from patients who died from suicide.
Human blood and other peripheral tissue gene expression data
For human blood gene expression, evidence was extracted from our database compiled by a similar method as above, performing a search of the primary literature by entering various combinations of keywords (gene name, suicide, suicide gene expression, lymphoblasts and blood). No matches were found for our final list of differentially expressed genes.
Human genetic evidence (association and linkage)
To designate convergence for a particular gene, the gene had to have independent published evidence of association or linkage for suicide. For linkage, the location of each gene was obtained through GeneCards (http://www.genecards.org), and the sex averaged cM location of the start of the gene was then obtained through http://compgen.rutgers.edu/mapinterpolator. For linkage convergence, the start of the gene had to map within 5 cM of the location of a marker linked to the disorder.
CFG scoring
For CFG analysis (Figure 2), two external cross-validating lines of evidence were weighted such that findings in human postmortem brain tissue, the target organ, were prioritized over genetic findings, by giving it twice as many points. Human brain expression evidence was given 4 points, whereas human genetic evidence was given a maximum of 2 points for association and 1 point for linkage. Each line of evidence was capped in such a way that any positive findings within that line of evidence result in maximum points, regardless of how many different studies support that single line of evidence, to avoid potential popularity biases.
In addition to our external score, we also prioritized genes based upon the initial differential expression analyses used to identify them. Probesets identified by differential expression analyses could receive a maximum of 6 points (1 or 3 points from intrasubject analyses, and 1 or 3 points from intersubject analyses).
Thus, the maximum possible total CFG score for each gene was 12 points (6 points for the internal score+6 points for the external score), with the internal and external evidence weighted equally. The scoring system was decided upon before the analysis. It has not escaped our attention that other ways of scoring the lines of evidence may give slightly different results in terms of prioritization, if not in terms of the list of genes per se. Nevertheless, we feel this simple scoring system provides a good separation of genes based on differential expression and on independent cross-validating evidence in the field (Figure 2).
Pathway analyses
IPA 9.0 (Ingenuity Systems, www.ingenuity.com, Redwood City, CA, USA) was used to analyze the biological roles, including top canonical pathways and diseases, of the candidate genes resulting from our work (Table 3 and Supplementary Table S4), as well as to identify genes in our data sets that are the target of existing drugs (Supplementary Table S5). Pathways were identified from the IPA library of canonical pathways that were most significantly associated with genes in our data set. The significance of the association between the data set and the canonical pathway was measured in two ways: (1) A ratio of the number of molecules from the data set that map to the pathway divided by the total number of molecules that map to the canonical pathway is displayed. (2) Fisher’s exact test was used to calculate a P‐value determining the probability that the association between the genes in the data set and the canonical pathway is explained by chance alone. We also conducted a Kyoto Encyclopedia of Genes and Genomes pathway analysis through the Partek Genomic Suites 6.6 software package.
Validation analyses
We imported the nine Affymetrix microarray data files from the suicide completers cohort as cel files into the Partek Genomic Suites 6.6 software package (Partek Incorporated). We then ran a RMA, background corrected with quantile normalization, and a median polish probeset summarization of all the chips from the discovery and validation cohort (24+9=33 chips), to obtain the normalized expression levels of all probesets for each chip. Partek normalizes expression data into a log base of 2 for visualization purposes. We non-log-transformed expression data by taking 2 to the power of the transformed expression value. We then used the non-log-transformed expression data to compare expression levels of biomarkers in the different groups (Figure 3). One-tail Student’s t-tests with unequal variance, one-way ANOVA and Bonferonni corrections were used for statistical comparisons.
Testing of biomarkers in suicide completers. (a) Upper: SAT1 (spermidine/spermine N1–acetyltransferase 1) expression is significantly increased (P=0.0057) in our discovery work between subjects with high suicidal ideation (SI) (mean=3413.37) and those reporting no SI (mean=2642.97). Our test cohort of suicide completers (mean=7171.51) showed significantly greater expression of SAT1 than both high SI (P=7.27e-07) and no SI (P=1.51e-07) groups from the discovery cohort. Lower: a suicide risk score was calculated by scoring the s.d. band a subject fell within as derived from the high SI discovery cohort, starting from the mean of the high-SI discovery cohort. A score of 0 indicates the subject falling between the means of the high SI and no SI subjects in the discovery cohort. A score of 1 means between the mean of the high SI and the first s.d. above it, score of 2 between the first and second s.d., score of 3 between the second and third s.d., and so on. Red line marks where the average SAT1 gene expression in high SI subjects would fall. (b) Upper: CD24 (CD24 molecule/small cell lung carcinoma cluster 4 antigen) expression was significantly decreased (P=0.0044) within the discovery cohort between subjects reporting high SI (mean=73.01) and no SI (mean=108.634). The test cohort of suicide completers (mean=71.61) was also significantly decreased (P=0.0031) when compared with subjects reporting no SI. Lower: suicide risk score defined as the s.d. band in which the subject expression fell below the mean of the high-SI discovery cohort. Red line marks where the average CD24 gene expression in high SI subjects would fall. (c) Testing of top candidate biomarkers for suicidality. Thirteen out of the 41 CFG top-scoring biomarkers from Figure 2b (32%) showed step-wise significant change from no SI to high SI, to the validation suicide completers group. Six out of them (15%) remained significant after strict Bonferroni correction for multiple comparisons. The top CFG scoring biomarker SAT1 remained the top biomarker after validation.
For live cohorts’ future hospitalization analyses in bipolar disorder and schizophrenia/schizoaffective, we similarly RMA normalized each cohort, before looking at biomarker levels in individual subjects. One-tail Student’s t-tests with equal variance were used for statistical comparisons. Receiver-operating characteristic curves were calculated using SPSS software for each of the four-dimensional analyses, predicting the state variable of hospitalizations due to suicidality.
Results
Discovery
We conducted whole-genome gene expression profiling in the blood samples from a longitudinally followed homogeneous cohort of male subjects with a major mood disorder (bipolar disorder) that predisposes to suicidality. One in three individuals with bipolar disorder attempt suicide during their lifetime.13 The samples were collected at repeated visits, 3–6 months apart. State information about SI was collected from a questionnaire administered at the time of each blood draw (Table 1). Out of 75 bipolar subjects (with a total of 174 visits) followed longitudinally in our study, there were 9 subjects that switched from a no SI (SI score of 0) to a high SI state (SI score of 2 and above) at different visits, which was our intended study group. We used a powerful intrasubject design to analyze data from these 9 subjects and their 24 visits. An intrasubject design factors out genetic variability, as well as some medications, lifestyle and demographic effects on gene expression, permitting identification of relevant signal with Ns as small as 1.14 An ancillary benefit of an intrasubject design may be accuracy/consistency of self-report of psychiatric symptoms (‘phene expression’), similar in rationale to the signal-detection benefits it provides in gene expression. We also used an overall intersubject case–case analysis, to identify genes differentially expressed in the blood in no SI states versus high SI states (Figure 1). The number of subjects that met our criteria and were analyzed is small, but comparable to those in human postmortem brain gene expression studies of suicide.15 We are indeed treating the blood samples as surrogate tissue for brains, with the caveat that they are not the real target organ. However, with the blood samples from live human subjects we have the advantages of in-vivo accessibility, better knowledge of the mental state at the time of collection, less technical artifacts and especially of being able to do powerful intrasubject analyses from visit to visit. We considered and differentially scored only the very top 0.1 and 5% of the gene expression probesets distributions, and also required overlap between the intrasubject and intersubject analyses of gene expression changes. Such a restrictive approach was used as a way of minimizing false positives, even at the risk of having false negatives (Figure 1c). For example, there were genes on each of the two lists, from intra- and intersubject analyses, that had clear prior evidence for involvement in suicidality, such as MT1E15 and GSK3B, respectively,16 but were not included in our subsequent analyses because they were not in the overlap.
We then used a CFG approach (Figure 2) to cross match the list of 246 overlapping top differentially expressed genes from the blood samples with other key lines of evidence (human postmortem brain data and human genetic data) implicating them in suicidality, as a way of identifying and prioritizing disease-relevant genomic biomarkers, extracting generalizable signal out of potential cohort-specific residual noise and genetic heterogeneity. We have built in our lab manually curated databases of the psychiatric genomic and proteomic literature to date, for use in CFG analyses.12, 17, 18, 19 The CFG approach is thus a de facto field-wide collaboration. We use in essence, in a Bayesian fashion, the whole body of knowledge in the field to leverage findings from our discovery data sets. Unlike our use of CFG in previous studies, for the current one we did not use any human peripheral tissue evidence from the literature, as there was none directly matching our genes, reflecting perhaps the dearth of peripheral gene expression work done so far on suicides, and the need for a study like ours. We also did not use animal model evidence, as there are to date no clear studies in animal models of self-harm or suicidality. SAT1 was the top-scoring blood biomarker, with the most extensive convergent evidence, increased in suicidal states identified by our work (that is, the top risk marker). CD24 (CD24 molecule/small cell lung carcinoma cluster 4 antigen) was the top blood biomarker decreased in suicidal states (that is, the top protective marker; Figure 2 and Table 2).
Testing in suicide completers
In order to know whether our findings relate to actual completed suicide, we then tested SAT1 levels in the blood samples from a heterogeneous cohort of nine consecutive male suicide completers obtained from the coroner’s office, with the following characteristics: we required that the cases included in our analysis had a postmortem interval from last observed alive under 24 h, and that they had committed suicide by means other than overdoses, which could alter gene expression. Remarkably, we found SAT1 gene expression levels to be elevated in nine out of nine (100%) subjects who committed suicide, that we tested. In each of the suicide completers, the increase in SAT1 was at least three s.d. above the average levels in high SI subjects, which constitutes a very stringent threshold for use as a predictive biomarker (Figure 3). We also examined other top candidate biomarkers for suicidality (Figure 3 and Supplementary Figure S3). Remarkably, 13 out of the 41 CFG top-scoring biomarkers from Figure 2b (32%) showed step-wise significant change from no SI to high SI, to the test suicide completers group. Six out of them (15%) remained significant after strict Bonferroni correction for multiple comparisons (Figure 3). The top CFG scoring biomarker SAT1 remained the top biomarker after validation.
Mechanistic understanding
Pathway analyses of our suicidality biomarker data identified among the top pathways the omega-3 docosahexaenoic acid signaling pathway. Low omega-3 levels have been correlated with increased suicidality in human epidemiological studies.20, 21 Several of the biomarkers from our current study (SAT1, S100A8, IL1B and 16 others) were changed in expression by omega-3 treatment in the blood of the circadian clock gene DBP (D-box binding protein) knock-out mouse model in opposite direction to our human suicidality data (Supplementary Table S6). DBP is also one of the biomarkers identified to be decreased in high suicidal states in the current analysis. Serendipitously, previous work by our group has implicated DBP in mood disorders,22 psychosis,23 alcoholism9 and anxiety disorders.10 Mice engineered to lack DBP were stress-reactive and displayed a behavioral phenotype similar to bipolar disorder and comorbid alcoholism.24 In addition to bipolar disorder, alcoholism increases risk for suicide.25 Phosphatase and tensin homolog (PTEN), a biomarker increased in suicidality in the current study in the blood, as well as in the brain of suicide completers,26 was also increased in the amygdala and was decreased in the prefrontal cortex of DBP knock-out mice subjected to stress.25 S100A8, another biomarker increased in suicidality in the current study, was also increased in the blood of DBP stressed mice. Treatment with omega-3 fatty acids normalized the phenotype of those mice.27
Other circadian clock-modulated genes identified by our analysis as biomarkers for suicidality were PIK3R5, MARCKS, IL1B, CASC1, CCRN4L, H3F3B, RBCK1, TNK2 and UBE2B. Circadian genes are involved in sleep–wake cycles, as well as mood regulation.6, 7, 22, 28, 29 Abnormal sleep (insomnia) has been identified as a risk factor for suicide.30 IL1B is also an inflammatory marker, and has previously been implicated by us in anxiety disorders.10
In addition, S100A8, MBNL2 and three other biomarkers had evidence for modulation by clozapine in the blood in opposite direction to our human suicidality data in previous independent animal model pharmacogenomics studies conducted by us4, 23 (Supplementary Table S6). Clozapine is the only FDA-approved treatment for suicidality.31
Thus, the convergent evidence for our biomarkers is strong in translational ways beyond those used for their discovery and selection. S100A8 may be a key biomarker to monitor in terms of response to treatment with classic (clozapine) and complementary (omega-3) agents. Other potential drugs to be studied for modulating suicidality were revealed by our analyses (Supplementary Tables S5 and S6).
SAT1, FOXN3, DISC1, MBNL2 and RHEB had genetic association evidence for suicidality, suggesting that they are not only state biomarkers but also trait factors influencing suicidal risk. DISC1 is also one of the top candidate genes for schizophrenia based on a large-scale CFG analysis of schizophrenia genome-wide association study we recently conducted,11 while DISC1 and MBNL2 are also among of the top candidate genes for bipolar disorder based on a large-scale CFG analysis of bipolar disorder genome-wide association study we previously conducted.7 In addition, DISC1 has clear animal model data for the role of its interaction with environmental stress in the pathophysiology of psychotic depression.32 DISC1 and MBNL2 may thus be key state and trait factors for suicidality risk in psychotic mood disorder subjects, and an indication for clozapine treatment in such subjects.
We also looked at the overlap of our suicide biomarkers with our previous mood biomarker3 and psychosis biomarker4 work (Supplementary Table S7), as well as with the human postmortem brain literature for other psychiatric disorders (Supplementary Table S8). DOCK5 and four other biomarkers were changed in high suicidal states in the opposite direction to their change in high mood states, and DOCK5 and six other biomarkers were changed in the same direction as their change in high psychosis states, suggesting that suicidality could be viewed as a psychotic dysphoric state, and that DOCK5 may be an additional key biomarker reflecting that state. This molecularly informed view is consistent with the emerging clinical evidence in the field.33
The convergence of evidence then suggests that at least in the population we studied, suicidality may be associated with dysphoric mood, as well as increased psychosis, anxiety and stress. In our own data, SAT1 blood gene expression levels showed a trend towards increase in low mood, high psychosis, high anxiety and high stress in our bipolar subjects (Figure 4).
SAT1 (spermidine/spermine N1–acetyltransferase 1) expression in the bipolar discovery cohort: relationship with suicidal ideation (SI), mood, psychosis, anxiety and stress. (a) SAT1 expression and SI item from Hamilton Rating Scale for Depression (HAMD) (scores of 0–4). (b) SAT1 expression and visual-analog scale for mood (0–100). High mood is to the left on the x-axis, low mood is to the right. (c) SAT1 expression and Hallucinations item from Positive and Negative Symptoms Scale (PANSS; scores of 1–7). Higher score indicates higher symptoms. (d) SAT1 expression and Delusions item from PANSS (scores of 1–7). Higher score indicates higher symptoms. (e) SAT1 expression and visual-analog scale for anxiety (0–100). Higher score indicates higher symptoms. (f) SAT1 expression and self-rating scale for stress (1–10). Higher score indicates higher symptoms. Only 20 out of 24 visits had stress data collected. *P<0.05 between highest symptoms and lowest symptoms group.
Prospective validation
To further validate SAT1, our top marker, we also looked at subsequent hospitalizations with and without suicidality (Table 1 and Supplementary Table S9), and previous hospitalizations with and without suicidality (Supplementary Table S10), in two live cohorts, one bipolar (n=42) and one psychosis (schizophrenia/schizoaffective; n=46). Higher SAT1 levels compared with lower SAT1 levels at the time of testing differentiated future and past hospitalizations owing to suicidality in the bipolar disorder subjects (Figure 5). A similar but weaker pattern was exhibited in the psychosis (schizophrenia/schizoaffective) subjects (Supplementary Figure S2). Remarkably, besides SAT1, three other (PTEN, MARCKS and MAP3K3) of the six biomarkers that survived Bonferroni correction in the suicide completers cohort validation step also showed similar but weaker results (Supplementary Table S11 and Supplementary Figure S3). Taken together, the prospective and retrospective hospitalization data suggests SAT1, PTEN, MARCKS and MAP3K3 might be not only state markers but perhaps trait markers as well.
Prospective validation of SAT1 (spermidine/spermine N1–acetyltransferase 1): follow-up of future psychiatric hospitalizations due to suicidality. We analyzed in 42 bipolar subjects whether their SAT1 levels at the time of initial testing differentiated those who had subsequent hospitalizations due to suicidality in the years since the testing occurred. Range was 0.33–5.92 years of follow-up, average 2.48 years. (a) Upper half of SAT1 scores versus lower half of SAT1 scores. Twenty-one subjects in each group. There were six psychiatric hospitalizations not due to suicidality, and eight psychiatric hospitalizations due to suicidality. (b) Upper tertile of SAT1 scores versus lower tertile of SAT1 scores. Fourteen subjects in each group. There were three psychiatric hospitalizations not due to suicidality, and four psychiatric hospitalizations due to suicidality.
We also examined whether using a multi-dimensional approach enhanced our ability to predict future hospitalizations, by adding data about mood, anxiety and psychosis to the data about the SAT1 expression levels (Figure 6). We found that the receiver-operating characteristic curve improved in a step-wise fashion, from an area under the curve of 0.640 with SAT1 alone, to an area under the curve of 0.798 with SAT1 and anxiety, area under the curve of 0.813 with SAT1, anxiety and mood, and area under the curve of 0.835 with SAT1, anxiety, mood and psychosis. From our preliminary work, we identified levels of SAT1 that provide different levels of sensitivity and specificity (Supplementary Table S12). The anxiety and mood information was obtained from simple visual-analog scales, previously described by us.34 The psychosis information is based on the combining of the scores on the hallucinations and delusions in the Positive and Negative Symptoms Scale (Supplementary Figure S5). Of note, this simple clinical–genomic approach does not directly ask about SI, which some individuals may deny or choose not to share with clinicians. Similar data were obtained for the panel of six top markers as shown in Supplementary Figure S6.
Multi-dimensional prediction of future psychiatric hospitalizations due to suicidality. We analyzed in 42 bipolar subjects whether their SAT1 (spermidine/spermine N1–acetyltransferase 1), anxiety, mood and psychosis levels at the time of initial testing differentiated from those who had subsequent hospitalizations due to suicidality in the years since the testing occurred. Data in each dimension was normalized to a 0–100 scale (with the mood visual-analog scale (VAS) inverted, as the assumption was made that depressed mood states would more closely correlate with suicidality). The angle between dimensions was assumed to be 90°, and a simple Pythagorean distance from origin score was calculated. The distribution of this score in the test cohort was used to generate a receiver-operating characteristic curve for hospitalizations due to suicidality. (a) ROC curve. (b) Detailed results. (c) Three-dimensional visualization.
Discussion
Using discovery in live subjects and validation in suicide completers, we found possible biomarkers for suicidality. Our top biomarker finding, SAT1, as well as PTEN, MARCKS and MAP3K3, were additionally validated by prospective and retrospective analyses in live subjects, looking at the ability to predict and differentiate future and past hospitalizations due to suicidality in bipolar disorder and psychosis (schizophrenia/schizoaffective; Supplementary Table S11).
Apoptosis
Beyond predictions, as a window into the biology of suicidality, the current work shows overlap at a gene and pathway level with apoptosis (Table 3, Supplementary Table S3 and S4). SAT1, for example, is a key catabolic enzyme for polyamines. Polyamine levels within cells control cell viability, and significant decreases in polyamine levels can result in apoptosis.35 They seem to reflect an endowment for cellular and organismal activity and growth, key characteristics of mood.3, 7, 36 SAT1, which is increased in live SI subjects and in suicide completers in our studies, is highly inducible by a variety of stimuli, including toxins, cytokines, heat shock, ischemia and other stresses. SAT1-overexpressing mice had alterations in their polyamine pool, hair loss, infertility and weight loss.37, 38 Turecki and colleagues39 have provided compelling evidence for changes in the polyamine system in the brain of suicide completers. CD24, our top biomarker decreased in suicidal subjects, also has roles in apoptosis. Mice lacking CD24 show an increased rate of apoptosis.40 It could be that simpler mechanisms related to cellular survival and programed cell-death decision have been recruited by evolution for higher mental functions, such as feelings, thoughts, actions and behaviors, leading to suicidality. In that sense, suicidality could be viewed as whole-organism apoptosis (‘self-poptosis’). Apoptosis mechanisms have previously been implicated in mood disorders, and their inhibition in affective resilience.41 Interestingly, lithium, a medication with clinical evidence for preventing suicidality in bipolar disorder,42 has anti-apoptotic effects at a cellular level.43 Imaging studies have shown reduced gray matter volume in the brain of individuals with bipolar disorder and history of suicide attempts. Long-term lithium treatment was associated with increased gray matter volumes in the same areas where suicide was associated with decreased gray matter.44
Conclusions and future directions
Taken together, our results have implications for the understanding of suicide, as well as for the development of objective laboratory tests and tools to track suicidal risk and response to treatment. First, our results open empirical avenues for future field trials, clinical testing and validation in various at-risk populations, including studies in individuals with major depressive disorder. The current work was based on subjects with bipolar disorder, psychosis (schizophrenia/schizoaffective disorder) and coroner’s office cases, which may represent a more externalizing or impulsive population and type of suicidality. Other types are likely to exist. Second, more work also needs to be done to examine potential gender and ethnicity differences. Our current work is based on male Caucasian subjects. Third, predicting suicidal feelings and thoughts (ideation) may be different than predicting suicidal actions and behaviors. Our current work has focused on suicide completers and hospitalizations, admittedly a more emergent concern. Fourth, state versus trait issues and sensitivity versus specificity for suicidality, for the individual markers identified by us, as well as for panels of markers and multi-modal approaches, need to be studied more extensively in different populations. Fifth, past individual and family history, as well as environmental context, may help improve predictive approaches. Our approach was very focused and reductionist, albeit with good results.
Given the fact that approximately one million people die of suicide worldwide each year, and this is a potentially preventable cause of death, the need for, urgency and importance of efforts such as ours cannot be overstated.
References
Nock MK, Borges G, Bromet EJ, Cha CB, Kessler RC, Lee S . Suicide and suicidal behavior. Epidemiol Rev 2008; 30: 133–154.
Berngruber TW, Lion S, Gandon S . Evolution of suicide as a defence strategy against pathogens in a spatially structured environment. Ecol Lett 2013; 16: 446–453.
Le-Niculescu H, Kurian SM, Yehyawi N, Dike C, Patel SD, Edenberg HJ et al. Identifying blood biomarkers for mood disorders using convergent functional genomics. Mol Psychiatr 2009; 14: 156–174.
Kurian SM, Le-Niculescu H, Patel SD, Bertram D, Davis J, Dike C et al. Identification of blood biomarkers for psychosis using convergent functional genomics. Mol Psychiatr 2011; 16: 37–58.
Niculescu A, Segal D, Kuczenski R, Barrett T, Hauger R, Kelsoe J . Identifying a series of candidate genes for mania and psychosis: a convergent functional genomics approach. Physiol Genomics 2000; 4: 83–91.
Ogden CA, Rich ME, Schork NJ, Paulus MP, Geyer MA, Lohr JB et al. Candidate genes, pathways and mechanisms for bipolar (manic-depressive) and related disorders: an expanded convergent functional genomics approach. Mol Psychiatr 2004; 9: 1007–1029.
Patel SD, Le-Niculescu H, Koller DL, Green SD, Lahiri DK, McMahon FJ et al. Coming to grips with complex disorders: genetic risk prediction in bipolar disorder using panels of genes identified through convergent functional genomics. Am J Med Genet B Neuropsychiatr Genet 2010; 153B: 850–877.
Le-Niculescu H, Patel SD, Bhat M, Kuczenski R, Faraone SV, Tsuang MT et al. Convergent functional genomics of genome-wide association data for bipolar disorder: comprehensive identification of candidate genes, pathways and mechanisms. Am J Med Genet B Neuropsychiatr Genet 2009; 150B: 155–181.
Rodd ZA, Bertsch BA, Strother WN, Le-Niculescu H, Balaraman Y, Hayden E et al. Candidate genes, pathways and mechanisms for alcoholism: an expanded convergent functional genomics approach. Pharmacogenomics J 2007; 7: 222–256.
Le-Niculescu H, Balaraman Y, Patel SD, Ayalew M, Gupta J, Kuczenski R et al. Convergent functional genomics of anxiety disorders: translational identification of genes, biomarkers, pathways and mechanisms. Transl Psychiatr 2011; 1: e9.
Ayalew M, Le-Niculescu H, Levey DF, Jain N, Changala B, Patel SD et al. Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol Psychiatr 2012; 17: 887–905.
Niculescu AB, Le-Niculescu H . Convergent Functional Genomics: what we have learned and can learn about genes, pathways, and mechanisms. Neuropsychopharmacology 2010; 35: 355–356.
Novick DM, Swartz HA, Frank E . Suicide attempts in bipolar I and bipolar II disorder: a review and meta-analysis of the evidence. Bipolar Disord 2010; 12: 1–9.
Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HY, Miriami E et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 2012; 148: 1293–1307.
Sequeira A, Morgan L, Walsh DM, Cartagena PM, Choudary P, Li J et al. Gene expression changes in the prefrontal cortex, anterior cingulate cortex and nucleus accumbens of mood disorders subjects that committed suicide. PloS One 2012; 7: e35367.
Karege F, Perroud N, Burkhardt S, Schwald M, Ballmann E, La Harpe R et al. Alteration in kinase activity but not in protein levels of protein kinase B and glycogen synthase kinase-3beta in ventral prefrontal cortex of depressed suicide victims. Biol Psychiatry 2007; 61: 240–245.
Bertsch B, Ogden CA, Sidhu K, Le-Niculescu H, Kuczenski R, Niculescu AB . Convergent functional genomics: a Bayesian candidate gene identification approach for complex disorders. Methods 2005; 37: 274–279.
Le-Niculescu H, McFarland MJ, Mamidipalli S, Ogden CA, Kuczenski R, Kurian SM et al. Convergent Functional Genomics of bipolar disorder: from animal model pharmacogenomics to human genetics and biomarkers. Neurosci Biobehav Rev 2007; 31: 897–903.
Le-Niculescu H, Patel SD, Niculescu AB . Convergent integration of animal model and human studies of bipolar disorder (manic-depressive illness). Curr Opin Pharmacol 2010; 10: 594–600.
Sublette ME, Hibbeln JR, Galfalvy H, Oquendo MA, Mann JJ . Omega-3 polyunsaturated essential fatty acid status as a predictor of future suicide risk. Am J Psychiatry 2006; 163: 1100–1102.
Lewis MD, Hibbeln JR, Johnson JE, Lin YH, Hyun DY, Loewke JD . Suicide deaths of active-duty US military and omega-3 fatty-acid status: a case-control comparison. J Clin Psychiatry 2011; 72: 1585–1590.
Niculescu AB 3rd, Segal DS, Kuczenski R, Barrett T, Hauger RL, Kelsoe JR . Identifying a series of candidate genes for mania and psychosis: a convergent functional genomics approach. Physiol Genomics 2000; 4: 83–91.
Le-Niculescu H, Balaraman Y, Patel S, Tan J, Sidhu K, Jerome RE et al. Towards understanding the schizophrenia code: an expanded convergent functional genomics approach. Am J Med Genet B Neuropsychiatr Genet 2007; 144B: 129–158.
Le-Niculescu H, McFarland MJ, Ogden CA, Balaraman Y, Patel S, Tan J et al. Phenomic, convergent functional genomic, and biomarker studies in a stress-reactive genetic animal model of bipolar disorder and co-morbid alcoholism. Am J Med Genet B Neuropsychiatr Genet 2008; 147B: 134–166.
Oquendo MA, Currier D, Liu SM, Hasin DS, Grant BF, Blanco C . Increased risk for suicidal behavior in comorbid bipolar disorder and alcohol use disorders: results from the National Epidemiologic Survey on Alcohol and Related Conditions (NESARC). J Clin Psychiatry 2010; 71: 902–909.
Dwivedi Y, Rizavi HS, Zhang H, Roberts RC, Conley RR, Pandey GN . Modulation in activation and expression of phosphatase and tensin homolog on chromosome ten, Akt1, and 3-phosphoinositide-dependent kinase 1: further evidence demonstrating altered phosphoinositide 3-kinase signaling in postmortem brain of suicide subjects. Biol Psychiatry 2010; 67: 1017–1025.
Le-Niculescu H, Case NJ, Hulvershorn L, Patel SD, Bowker D, Gupta J et al. Convergent functional genomic studies of omega-3 fatty acids in stress reactivity, bipolar disorder and alcoholism. Transl Psychiatry 2011; 1: e4.
McGrath CL, Glatt SJ, Sklar P, Le-Niculescu H, Kuczenski R, Doyle AE et al. Evidence for genetic association of RORB with bipolar disorder. BMC Psychiatry 2009; 9: 70.
Bunney BG, Bunney WE . Mechanisms of Rapid Antidepressant Effects of Sleep Deprivation Therapy: Clock Genes and Circadian Rhythms. Biol Psychiatry 2012; 73: 1164–1171.
McCall WV, Batson N, Webster M, Case LD, Joshi I, Derreberry T et al. Nightmares and Dysfunctional Beliefs about Sleep Mediate the Effect of Insomnia Symptoms on Suicidal Ideation. J Clin Sleep Med 2013; 9: 135–140.
Meltzer HY, Alphs L, Green AI, Altamura AC, Anand R, Bertoldi A et al. Clozapine treatment for suicidality in schizophrenia: International Suicide Prevention Trial (InterSePT). Arch Gen Psychiatry 2003; 60: 82–91.
Niwa M, Jaaro-Peled H, Tankou S, Seshadri S, Hikida T, Matsumoto Y et al. Adolescent stress-induced epigenetic control of dopaminergic neurons via glucocorticoids. Science 2013; 339: 335–339.
Kelleher I, Lynch F, Harley M, Molloy C, Roddy S, Fitzpatrick C et al. Psychotic symptoms in adolescence inex risk for suicidal behavior: findings from 2 population-based case-control clinical interview studies. Arch Gen Psychiatry 2012; 69: 1277–1283.
Niculescu AB, Lulow LL, Ogden CA, Le-Niculescu H, Salomon DR, Schork NJ et al. PhenoChipping of psychotic disorders: a novel approach for deconstructing and quantitating psychiatric phenotypes. Am J Med Genet B Neuropsychiatr Genet 2006; 141B: 653–662.
Schipper RG, Penning LC, Verhofstad AA . Involvement of polyamines in apoptosis. Facts and controversies: effectors or protectors? Semin Cancer Biol 2000; 10: 55–68.
Niculescu AB 3rd, Schork NJ, Salomon DR . Mindscape: a convergent perspective on life, mind, consciousness and happiness. J Affect Disord 2010; 123: 1–8.
Pietila M, Alhonen L, Halmekyto M, Kanter P, Janne J, Porter CW . Activation of polyamine catabolism profoundly alters tissue polyamine pools and affects hair growth and female fertility in transgenic mice overexpressing spermidine/spermine N1-acetyltransferase. J Biol Chem 1997; 272: 18746–18751.
Min SH, Simmen RC, Alhonen L, Halmekyto M, Porter CW, Janne J et al. Altered levels of growth-related and novel gene transcripts in reproductive and other tissues of female mice overexpressing spermidine/spermine N1-acetyltransferase (SSAT). J Biol Chem 2002; 277: 3647–3657.
Fiori LM, Bureau A, Labbe A, Croteau J, Noel S, Merette C et al. Global gene expression profiling of the polyamine system in suicide completers. Int J Neuropsychopharmacol 2011; 14: 595–605.
Duckworth CA, Clyde D, Pritchard DM . CD24 is expressed in gastric parietal cells and regulates apoptosis and the response to Helicobacter felis infection in the murine stomach. Am J Physioly Gastrointest Liver Physiol 2012; 303: G915–G926.
Malkesman O, Austin DR, Tragon T, Henter ID, Reed JC, Pellecchia M et al. Targeting the BH3-interacting domain death agonist to develop mechanistically unique antidepressants. Mol Psychiatry 2012; 17: 770–780.
Tondo L, Baldessarini RJ . Long-term lithium treatment in the prevention of suicidal behavior in bipolar disorder patients. Epidemiol Psichiatr Soc 2009; 18: 179–183.
Lowthert L, Leffert J, Lin A, Umlauf S, Maloney K, Muralidharan A et al. Increased ratio of anti-apoptotic to pro-apoptotic Bcl2 gene-family members in lithium-responders one month after treatment initiation. Biol Mood Anxiety Disord 2012; 2: 15.
Benedetti F, Radaelli D, Poletti S, Locatelli C, Falini A, Colombo C et al. Opposite effects of suicidality and lithium on gray matter volumes in bipolar depression. J Affect Disord 2011; 135: 139–147.
Fiori LM, Wanner B, Jomphe V, Croteau J, Vitaro F, Tremblay RE et al. Association of polyaminergic loci with anxiety, mood disorders, and attempted suicide. PloS One 2010; 5: e15146.
Sequeira A, Gwadry FG, Ffrench-Mullen JM, Canetti L, Gingras Y, Casero RA Jr. et al. Implication of SSAT by gene expression and genetic variation in suicide and major depression. Arch Gen Psychiatry 2006; 63: 35–48.
Chen GG, Fiori LM, Moquin L, Gratton A, Mamer O, Mechawar N et al. Evidence of altered polyamine concentrations in cerebral cortex of suicide completers. Neuropsychopharmacology 2010; 35: 1477–1484.
Fiori LM, Turecki G . Epigenetic regulation of spermidine/spermine N1-acetyltransferase (SAT1) in suicide. J Psychiatr Res 2011; 45: 1229–1235.
Fiori LM, Mechawar N, Turecki G . Identification and characterization of spermidine/spermine N1-acetyltransferase promoter variants in suicide completers. Biol Psychiatry 2009; 66: 460–467.
Fiori LM, Zouk H, Himmelman C, Turecki G . X chromosome and suicide. Mol Psychiatry 2011; 16: 216–226.
Guipponi M, Deutsch S, Kohler K, Perroud N, Le Gal F, Vessaz M et al. Genetic and epigenetic analysis of SSAT gene dysregulation in suicidal behavior. Am J Med Genet B Neuropsychiatr Genet 2009; 150B: 799–807.
Klempan TA, Rujescu D, Merette C, Himmelman C, Sequeira A, Canetti L et al. Profiling brain expression of the spermidine/spermine N1-acetyltransferase 1 (SAT1) gene in suicide. Am J Med Genet B Neuropsychiatr Genet 2009; 150B: 934–943.
Galfalvy H, Zalsman G, Huang YY, Murphy L, Rosoklija G, Dwork AJ et al. A pilot genome wide association and gene expression array study of suicide with and without major depression. World J Biol Psychiatry 2011 (e-pub ahead of print).
Kekesi KA, Juhasz G, Simor A, Gulyassy P, Szego EM, Hunyadi-Gulyas E et al. Altered functional protein networks in the prefrontal cortex and amygdala of victims of suicide. PLoS One 2012; 7: e50532.
Kim S, Choi KH, Baykiz AF, Gershenfeld HK . Suicide candidate genes associated with bipolar disorder and schizophrenia: an exploratory gene expression profiling analysis of post-mortem prefrontal cortex. BMC Genomics 2007; 8: 413.
Klempan TA, Sequeira A, Canetti L, Lalovic A, Ernst C, ffrench-Mullen J et al. Altered expression of genes involved in ATP biosynthesis and GABAergic neurotransmission in the ventral prefrontal cortex of suicides with and without major depression. Mol Psychiatry 2009; 14: 175–189.
Pandey GN, Rizavi HS, Ren X, Fareed J, Hoppensteadt DA, Roberts RC et al. Proinflammatory cytokines in the prefrontal cortex of teenage suicide victims. J Psychiatr Res 2012; 46: 57–63.
Lalovic A, Klempan T, Sequeira A, Luheshi G, Turecki G . Altered expression of lipid metabolism and immune response genes in the frontal cortex of suicide completers. J Affect Disord 2010; 120: 24–31.
Sequeira A, Klempan T, Canetti L, ffrench-Mullen J, Benkelfat C, Rouleau GA et al. Patterns of gene expression in the limbic system of suicides with and without major depression. Mol Psychiatry 2007; 12: 640–655.
Zhurov V, Stead JD, Merali Z, Palkovits M, Faludi G, Schild-Poulter C et al. Molecular pathway reconstruction and analysis of disturbed gene expression in depressed individuals who died by suicide. PLoS One 2012; 7: e47581.
Freemantle E, Mechawar N, Turecki G . Cholesterol and phospholipids in frontal cortex and synaptosomes of suicide completers: relationship with endosomal lipid trafficking genes. J Psychiatr Res 2013; 47: 272–279.
Pandey GN, Dwivedi Y, Ren X, Rizavi HS, Roberts RC, Conley RR et al. Altered expression and phosphorylation of myristoylated alanine-rich C kinase substrate (MARCKS) in postmortem brain of suicide victims with or without depression. J Psychiatr Res 2003; 37: 421–432.
Menke A, Domschke K, Czamara D, Klengel T, Hennings J, Lucae S et al. Genome-wide association study of antidepressant treatment-emergent suicidal ideation. Neuropsychopharmacology 2012; 37: 797–807.
Butler AW, Breen G, Tozzi F, Craddock N, Gill M, Korszun A et al. A genomewide linkage study on suicidality in major depressive disorder confirms evidence for linkage to 2p12. Am J Med Genet B Neuropsychiatr Genet 2010; 153B: 1465–1473.
Acknowledgements
This work is, in essence, a field-wide collaboration. We would like to acknowledge our debt of gratitude for the efforts and results of the many other groups, cited in our paper, who have conducted and published studies (genetic and gene expression) in suicidality. With their arduous and careful work, a convergent approach, such as ours, is possible. We would particularly like to thank the veterans and other subjects who volunteered to participate in these studies, their families and their caregivers. Without their generous contribution, such work to advance the understanding of mental illness and help others would not be possible. We would like to thank Terri Gelbart for excellent technical help on the microarray work, and Dawn Graham for help with the human subjects data. This work was supported by an NIH Directors’ New Innovator Award (1DP2OD007363) and a VA Merit Award (1I01CX000139-01) to ABN.
Author Contributions
ABN designed the study and wrote the manuscript. HLN, DFL and MA analyzed the data. LP, LMG, NJ, EW, SB and GS performed database work. EB, KO, HD, JV, RS and MR organized and conducted testing in bipolar disorder subjects. MY, AB, AS and GES organized and carried out postmortem sample collections. NJS, SMK and DRS conducted microarray experiments and provided input on data analyses. All authors discussed the results and commented on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Molecular Psychiatry website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Le-Niculescu, H., Levey, D., Ayalew, M. et al. Discovery and validation of blood biomarkers for suicidality. Mol Psychiatry 18, 1249–1264 (2013). https://doi.org/10.1038/mp.2013.95
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mp.2013.95
Keywords
This article is cited by
-
Towards precision medicine for anxiety disorders: objective assessment, risk prediction, pharmacogenomics, and repurposed drugs
Molecular Psychiatry (2023)
-
Proteins associated with future suicide attempts in bipolar disorder: A large-scale biomarker discovery study
Molecular Psychiatry (2022)
-
Identification of potential blood biomarkers associated with suicide in major depressive disorder
Translational Psychiatry (2022)
-
Cerebrospinal fluid concentration of complement component 4A is increased in first episode schizophrenia
Nature Communications (2022)
-
Associations of serum cystatin C with depressive symptoms and suicidal ideation in major depressive disorder
BMC Psychiatry (2021)