Abstract
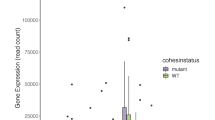
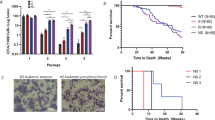
Acute myelogenous leukemia (AML) is a high-risk hematopoietic malignancy caused by a variety of mutations, including genes encoding the cohesin complex. Recent studies have demonstrated that reduction in cohesin complex levels leads to enhanced self-renewal in hematopoietic stem and progenitors (HSPCs). We sought to delineate the molecular mechanisms by which cohesin mutations promote enhanced HSPC self-renewal as this represents a critical initial step during leukemic transformation. We verified that RNAi against the cohesin subunit Rad21 causes enhanced self-renewal of HSPCs in vitro through derepression of polycomb repressive complex 2 (PRC2) target genes, including Hoxa7 and Hoxa9. Importantly, knockdown of either Hoxa7 or Hoxa9 suppressed self-renewal, implying that both are critical downstream effectors of reduced cohesin levels. We further demonstrate that the cohesin and PRC2 complexes interact and are bound in close proximity to Hoxa7 and Hoxa9. Rad21 depletion resulted in decreased levels of H3K27me3 at the Hoxa7 and Hoxa9 promoters, consistent with Rad21 being critical to proper gene silencing by recruiting the PRC2 complex. Our data demonstrates that the cohesin complex regulates PRC2 targeting to silence Hoxa7 and Hoxa9 and negatively regulate self-renewal. Our studies identify a novel epigenetic mechanism underlying leukemogenesis in AML patients with cohesin mutations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Burnett A, Wetzler M, Lowenberg B . Therapeutic advances in acute myeloid leukemia. J Clin Oncol 2011; 29: 487–494.
Thakar MS, Talano J-AM, Tower RL, Kelly ME, Burke MJ . Indications for transplantation in childhood acute leukemia and the impact of minimal residual disease on relapse: a review. Clin Pract 2014; 11: 79–90.
Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013; 368: 2059–2074.
Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 2012; 150: 264–278.
Kon A, Shih L-Y, Minamino M, Sanada M, Shiraishi Y, Nagata Y et al. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet 2013; 45: 1232–1237.
Thota S, Viny AD, Makishima H, Spitzer B, Radivoyevitch T, Przychodzen B et al. Genetic alterations of the cohesin complex genes in myeloid malignancies. Blood 2014; 124: 1790–1798.
Thol F, Bollin R, Gehlhaar M, Walter C, Dugas M, Suchanek KJ et al. Mutations in the cohesin complex in acute myeloid leukemia: clinical and prognostic implications. Blood 2014; 123: 914–920.
Garg M, Nagata Y, Kanojia D, Mayakonda A, Yoshida K, Haridas Keloth S et al. Profiling of somatic mutations in acute myeloid leukemia with FLT3-ITD at diagnosis and relapse. Blood 2015; 126: 2491–2501.
Xu H, Balakrishnan K, Malaterre J, Beasley M, Yan Y, Essers J et al. Rad21-cohesin haploinsufficiency impedes DNA repair and enhances gastrointestinal radiosensitivity in mice. PLoS One 2010; 5: e12112.
Remeseiro S, Cuadrado A, Gómez-López G, Pisano DG, Losada A . A unique role of cohesin-SA1 in gene regulation and development. EMBO J 2012; 31: 2090–2102.
Kawauchi S, Calof AL, Santos R, Lopez-Burks ME, Young CM, Hoang MP et al. Multiple organ system defects and transcriptional dysregulation in the Nipbl+/− mouse, a model of Cornelia de Lange syndrome. PLoS Genet 2009; 5: e1000650.
White JK, Gerdin A-K, Karp NA, Ryder E, Buljan M, Bussell JN et al. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell 2013; 154: 452–464.
Leeke B, Marsman J, O’Sullivan JM, Horsfield JA . Cohesin mutations in myeloid malignancies: underlying mechanisms. Exp Hematol Oncol 2014; 3: 13.
Yoshida K, Toki T, Okuno Y, Kanezaki R, Shiraishi Y, Sato-Otsubo A et al. The landscape of somatic mutations in Down syndrome-related myeloid disorders. Nat Genet 2013; 45: 1293–1299.
Mehta GD, Kumar R, Srivastava S, Ghosh SK . Cohesin: functions beyond sister chromatid cohesion. FEBS Lett 2013; 587: 2299–2312.
Williams MS, Somervaille TCP . Leukemogenic activity of cohesin rings true. Cell Stem Cell 2015; 17: 642–644.
Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 2008; 451: 796–801.
Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 2010; 467: 430–435.
Merkenschlager M, Odom DT . CTCF and cohesin: linking gene regulatory elements with their targets. Cell 2013; 152: 1285–1297.
Baranello L, Kouzine F, Levens D . CTCF and cohesin cooperate to organize the 3D structure of the mammalian genome. Proc Natl Acad Sci USA 2014; 111: 889–890.
Xu M, Zhao G-N, Lv X, Liu G, Wang LY, Hao D-L et al. CTCF controls HOXA cluster silencing and mediates PRC2-repressive higher-order chromatin structure in NT2/D1 cells. Mol Cell Biol 2014; 34: 3867–3879.
Li T, Hu J-F, Qiu X, Ling J, Chen H, Wang S et al. CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex 2 intrachromosomal loop. Mol Cell Biol 2008; 28: 6473–6482.
Mazumdar C, Shen Y, Xavy S, Zhao F, Reinisch A, Li R et al. Leukemia-associated cohesin mutants dominantly enforce stem cell programs and impair human hematopoietic progenitor differentiation. Cell Stem Cell 2015; 17: 675–688.
Viny AD, Ott CJ, Spitzer B, Rivas M, Meydan C, Papalexi E et al. Dose-dependent role of the cohesin complex in normal and malignant hematopoiesis. J Exp Med 2015; 212: 1819–1832.
Mullenders J, Aranda-Orgilles B, Lhoumaud P, Keller M, Pae J, Wang K et al. Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. J Exp Med 2015; 212: 1833–1850.
Galeev R, Baudet A, Kumar P, Rundberg Nilsson A, Nilsson B, Soneji S et al. Genome-wide RNAi screen identifies cohesin genes as modifiers of renewal and differentiation in human HSCs. Cell Rep 2016; 14: 2988–3000.
Castronovo P, Gervasini C, Cereda A, Masciadri M, Milani D, Russo S et al. Premature chromatid separation is not a useful diagnostic marker for Cornelia de Lange syndrome. Chromosome Res 2009; 17: 763–771.
Liu J, Krantz ID . Cornelia de Lange syndrome, cohesin, and beyond. Clin Genet 2009; 76: 303–314.
Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 1999; 286: 531–537.
Spencer DH, Young MA, Lamprecht TL, Helton NM, Fulton R, O'Laughlin M et al. Epigenomic analysis of the HOX gene loci reveals mechanisms that may control canonical expression patterns in AML and normal hematopoietic cells. Leukemia 2015; 29: 1279–1289.
Li Z, Zhang Z, Li Y, Arnovitz S, Chen P, Huang H et al. PBX3 is an important cofactor of HOXA9 in leukemogenesis. Blood 2013; 121: 1422–1431.
Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G . Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J 1998; 17: 3714–3725.
Thorsteinsdottir U, Mamo A, Kroon E, Jerome L, Bijl J, Lawrence HJ et al. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood 2002; 99: 121–129.
Muntean AG, Tan J, Sitwala K, Huang Y, Bronstein J, Connelly JA et al. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell 2010; 17: 609–621.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550.
Lawrence HJ, Rozenfeld S, Cruz C, Matsukuma K . Frequent co-expression of the HOXA9 and MEIS1 homeobox genes in human myeloid leukemias. Leukemia 1999; 13: 1993–1999.
Afonja O, Smith JE, Cheng DM, Goldenberg AS, Amorosi E, Shimamoto T et al. MEIS1 and HOXA7 genes in human acute myeloid leukemia. Leuk Res 2000; 24: 849–855.
Thorsteinsdottir U, Kroon E, Jerome L, Blasi F, Sauvageau G . Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol Cell Biol 2001; 21: 224–234.
Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K . Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 2006; 20: 1123–1136.
Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE et al. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature 2011; 476: 467–471.
Seitan VC, Faure AJ, Zhan Y, McCord RP, Lajoie BR, Ing-Simmons E et al. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res 2013; 23: 2066–2077.
Ayton PM . Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev 2003; 17: 2298–2307.
Kroon E, Thorsteinsdottir U, Mayotte N, Nakamura T, Sauvageau G . NUP98–HOXA9 expression in hemopoietic stem cells induces chronic and acute myeloid leukemias in mice. EMBO J 2001; 20: 350–361.
Faber J, Krivtsov AV, Stubbs MC, Wright R, Davis TN, van den Heuvel-Eibrink M et al. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood 2009; 113: 2375–2385.
Nagel S, Venturini L, Marquez VE, Meyer C, Kaufmann M, Scherr M et al. Polycomb repressor complex 2 regulates HOXA9 and HOXA10, activating ID2 in NK/T-cell lines. Mol Cancer 2010; 9: 151.
Sauvageau G, Lansdorp PM, Eaves CJ, Hogge DE, Dragowska WH, Reid DS et al. Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proc Natl Acad Sci USA 1994; 91: 12223–12227.
Lawrence HJ . Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood 2005; 106: 3988–3994.
Kamminga LM, Bystrykh LV, de Boer A, Houwer S, Douma J, Weersing E et al. The Polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood 2006; 107: 2170–2179.
Cao Q, Wang X, Zhao M, Yang R, Malik R, Qiao Y et al. The central role of EED in the orchestration of polycomb group complexes. Nat Commun 2014; 5: 3127.
Di Croce L, Helin K . Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol 2013; 20: 1147–1155.
Simon JA, Kingston RE . Occupying chromatin: polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell 2013; 49: 808–824.
Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012; 485: 376–380.
Sanyal A, Lajoie BR, Jain G, Dekker J . The long-range interaction landscape of gene promoters. Nature 2013; 489: 109–113.
Ong C-T, Corces VG . CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet 2014; 15: 234–246.
Phillips JE, Corces VG . CTCF: master weaver of the genome. Cell 2009; 137: 1194–1211.
Berlivet S, Paquette D, Dumouchel A, Langlais D, Dostie J, Kmita M . Clustering of tissue-specific sub-TADs accompanies the regulation of HoxA genes in developing limbs. PLoS Genet 2013; 9: e1004018.
Vieux-Rochas M, Fabre PJ, Leleu M, Duboule D, Noordermeer D . Clustering of mammalian Hox genes with other H3K27me3 targets within an active nuclear domain. Proc Natl Acad Sci 2015; 112: 4672–4677.
Noordermeer D, Leleu M, Schorderet P, Joye E, Chabaud F, Duboule D . Temporal dynamics and developmental memory of 3D chromatin architecture at Hox gene loci. Elife 2014; 3: e02557.
Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 2011; 20: 66–78.
Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM et al. hDOT1L links histone methylation to leukemogenesis. Cell 2005; 121: 167–178.
Chen S, Yang Z, Wilkinson AW, Deshpande AJ, Sidoli S, Krajewski K et al. The PZP domain of AF10 senses unmodified H3K27 to regulate DOT1L-mediated methylation of H3K79. Mol Cell 2015; 60: 319–327.
Acknowledgements
We thank Hope Campbell and Benedetta Bonacci for their assistance with flow cytometry. S Rao is supported by grants from the Midwest Athletes Against Childhood Cancer (MACC Fund), Hyundai Hope on Wheels and an American Society of Hematology Bridge Grant. Additional research support to S Rao was provided by charitable gifts from Hartland Blood Centers, Ms Nanette Gardetto and Mr Doug Ziegler.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Rights and permissions
About this article
Cite this article
Fisher, J., Peterson, J., Reimer, M. et al. The cohesin subunit Rad21 is a negative regulator of hematopoietic self-renewal through epigenetic repression of Hoxa7 and Hoxa9. Leukemia 31, 712–719 (2017). https://doi.org/10.1038/leu.2016.240
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2016.240
This article is cited by
-
Three-dimensional genome organization in immune cell fate and function
Nature Reviews Immunology (2023)
-
Combinatorial genetics reveals the Dock1-Rac2 axis as a potential target for the treatment of NPM1;Cohesin mutated AML
Leukemia (2022)
-
DOT1L inhibitors block abnormal self-renewal induced by cohesin loss
Scientific Reports (2021)
-
High CENPM mRNA expression and its prognostic significance in hepatocellular carcinoma: a study based on data mining
Cancer Cell International (2020)
-
Chronic loss of STAG2 leads to altered chromatin structure contributing to de-regulated transcription in AML
Journal of Translational Medicine (2020)