Abstract
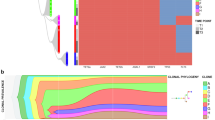
Clonal architecture in myeloproliferative neoplasms (MPNs) is poorly understood. Here we report genomic analyses of a patient with primary myelofibrosis (PMF) transformed to secondary acute myeloid leukemia (sAML). Whole genome sequencing (WGS) was performed on PMF and sAML diagnosis samples, with skin included as a germline surrogate. Deep sequencing validation was performed on the WGS samples and an additional sample obtained during sAML remission/relapsed PMF. Clustering analysis of 649 validated somatic single-nucleotide variants revealed four distinct clonal groups, each including putative driver mutations. The first group (including JAK2 and U2AF1), representing the founding clone, included mutations with high frequency at all three disease stages. The second clonal group (including MYB) was present only in PMF, suggesting the presence of a clone that was dispensable for transformation. The third group (including ASXL1) contained mutations with low frequency in PMF and high frequency in subsequent samples, indicating evolution of the dominant clone with disease progression. The fourth clonal group (including IDH1 and RUNX1) was acquired at sAML transformation and was predominantly absent at sAML remission/relapsed PMF. Taken together, these findings illustrate the complex clonal dynamics associated with disease evolution in MPNs and sAML.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Levine RL, Gilliland DG . Myeloproliferative disorders. Blood 2008; 112: 2190–2198.
Tefferi A, Vainchenker W . Myeloproliferative neoplasms: molecular pathophysiology, essential clinical understanding, and treatment strategies. J Clin Oncol 2011; 29: 573–582.
Gangat N, Caramazza D, Vaidya R, George G, Begna K, Schwager S et al. DIPSS plus: a refined Dynamic International Prognostic Scoring System for primary myelofibrosis that incorporates prognostic information from karyotype, platelet count, and transfusion status. J Clin Oncol 2011; 29: 392–397.
Heaney ML, Soriano G . Acute myeloid leukemia following a myeloproliferative neoplasm: clinical characteristics, genetic features and effects of therapy. Curr Hematol Malig Rep 2013; 8: 116–122.
Mesa RA, Li CY, Ketterling RP, Schroeder GS, Knudson RA, Tefferi A . Leukemic transformation in myelofibrosis with myeloid metaplasia: a single-institution experience with 91 cases. Blood 2005; 105: 973–977.
Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 2005; 365: 1054–1061.
James C, Ugo V, Le Couedic JP, Staerk J, Delhommeau F, Lacout C et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 2005; 434: 1144–1148.
Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med 2005; 352: 1779–1790.
Levine R, Wadleigh M, Cools J, Ebert B, Wernig G, Huntly BJ et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 2005; 7: 387–397.
Pardanani A, Levine R, Lasho T, Pikman Y, Mesa R, Wadleigh M et al. MPL515 mutations in myeloproliferative and other myeloid disorders: a study of 1182 patients. Blood 2006; 108: 3472–3476.
Pikman Y, Lee BH, Mercher T, McDowell E, Ebert B, Gozo M et al. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med 2006; 3: e270.
Oh ST, Simonds EF, Jones C, Hale MB, Goltsev Y, Gibbs KD Jr. et al. Novel mutations in the inhibitory adaptor protein LNK drive JAK-STAT signaling in patients with myeloproliferative neoplasms. Blood 2010; 116: 988–992.
Pardanani A, Lasho T, Finke C, Oh ST, Gotlib J, Tefferi A . LNK mutation studies in blast-phase myeloproliferative neoplasms, and in chronic-phase disease with TET2, IDH, JAK2 or MPL mutations. Leukemia 2010; 24: 1713–1718.
Verstovsek S, Mesa RA, Gotlib J, Levy RS, Gupta V, DiPersio JF et al. A double-blind, placebo-controlled trial of ruxolitinib for myelofibrosis. N Engl J Med 2012; 366: 799–807.
Harrison C, Kiladjian JJ, Al-Ali HK, Gisslinger H, Waltzman R, Stalbovskaya V et al. JAK inhibition with ruxolitinib versus best available therapy for myelofibrosis. N Engl J Med 2012; 366: 787–798.
Tefferi A . Mutations galore in myeloproliferative neoplasms: would the real Spartacus please stand up? Leukemia 2011; 25: 1059–1063.
Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A et al. Mutation in TET2 in myeloid cancers. N Engl J Med 2009; 360: 2289–2301.
Wang L, Swierczek SI, Drummond J, Hickman K, Kim SJ, Walker K et al. Whole-exome sequencing of polycythemia vera revealed novel driver genes and somatic mutation shared by T cells and granulocytes. Leukemia 2014; 28: 935–938.
Theocharides A, Boissinot M, Girodon F, Garand R, Teo S, Lippert E et al. Leukemic blasts in transformed JAK2-V617F-positive myeloproliferative disorders are frequently negative for the JAK2-V617F mutation. Blood 2007; 110: 375–379.
Abdel-Wahab O . Genetics of the myeloproliferative neoplasms. Curr Opin Hematol 2011; 18: 117–123.
Klampfl T, Gisslinger H, Harutyunyan AS, Nivarthi H, Rumi E, Milosevic JD et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. N Engl J Med 2013; 369: 2379–2390.
Nangalia J, Massie CE, Baxter EJ, Nice FL, Gundem G, Wedge DC et al. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N Engl J Med 2013; 369: 2391–2405.
Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 2012; 150: 264–278.
Cancer Genome Atlas Research N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013; 368: 2059–2074.
Wang L, Swierczek SI, Lanikova L, Kim SJ, Hickman K, Walker K et al. The relationship of JAK2(V617F) and acquired UPD at chromosome 9p in polycythemia vera. Leukemia 2014; 28: 938–941.
Wang X, LeBlanc A, Gruenstein S, Xu M, Mascarenhas J, Panzera B et al. Clonal analyses define the relationships between chromosomal abnormalities and JAK2V617F in patients with Ph-negative myeloproliferative neoplasms. Exp Hematol 2009; 37: 1194–1200.
Godfrey AL, Chen E, Pagano F, Silber Y, Campbell PJ, Green AR . Clonal analyses reveal associations of JAK2V617F homozygosity with hematologic features, age and gender in polycythemia vera and essential thrombocythemia. Haematologica 2013; 98: 718–721.
Lundberg P, Karow A, Nienhold R, Looser R, Hao-Shen H, Nissen I et al. Clonal evolution and clinical correlates of somatic mutations in myeloproliferative neoplasms. Blood 2014; 123: 2220–2228.
Merker JD, Roskin KM, Ng D, Pan C, Fisk DG, King JJ et al. Comprehensive whole-genome sequencing of an early-stage primary myelofibrosis patient defines low mutational burden and non-recurrent candidate genes. Haematologica 2013; 98: 1689–1696.
Li H, Durbin R . Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–1760.
Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001; 29: 308–311.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25: 2078–2079.
Ye K, Schulz MH, Long Q, Apweiler R, Ning Z . Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 2009; 25: 2865–2871.
Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 2009; 361: 1058–1066.
Venkatraman ES, Olshen AB . A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics 2007; 23: 657–663.
Trapnell C, Pachter L, Salzberg SL . TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009; 25: 1105–1111.
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010; 28: 511–515.
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B . Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 2008; 5: 621–628.
Chen K, Wallis JW, Kandoth C, Kalicki-Veizer JM, Mungall KL, Mungall AJ et al. BreakFusion: targeted assembly-based identification of gene fusions in whole transcriptome paired-end sequencing data. Bioinformatics 2012; 28: 1923–1924.
Larson DE, Harris CC, Chen K, Koboldt DC, Abbott TE, Dooling DJ et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics 2012; 28: 311–317.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303.
Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012; 22: 568–576.
Walter MJ, Shen D, Ding L, Shao J, Koboldt DC, Chen K et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med 2012; 366: 1090–1098.
Ester M, Kriegel HP, Sander J, Xu X . A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise. Proceedings of 2nd International Conference on Knowledge Discovery and Data Mining; 2–4 August 1996; Portland, OR. AAAI Press: Menlo Park, CA, 1996, pp 226–231.
Fraley C, Raftery A . Model-based clustering, discriminant analysis and density estimation. J Am Stat Assoc 2002; 97: 611–631.
Levine RL, Belisle C, Wadleigh M, Zahrieh D, Lee S, Chagnon P et al. X-inactivation-based clonality analysis and quantitative JAK2V617F assessment reveal a strong association between clonality and JAK2V617F in PV but not ET/MMM, and identifies a subset of JAK2V617F-negative ET and MMM patients with clonal hematopoiesis. Blood 2006; 107: 4139–4141.
Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 2012; 481: 506–510.
Oh ST, Gotlib J . JAK2 V617F and beyond: role of genetics and aberrant signaling in the pathogenesis of myeloproliferative neoplasms. Expert Rev Hematol 2010; 3: 323–337.
Zhang SJ, Rampal R, Manshouri T, Patel J, Mensah N, Kayserian A et al. Genetic analysis of patients with leukemic transformation of myeloproliferative neoplasms shows recurrent SRSF2 mutations that are associated with adverse outcome. Blood 2012; 119: 4480–4485.
Steensma DP, Pardanani A, Stevenson WS, Hoyt R, Kiu H, Grigg AP et al. More on Myb in myelofibrosis: molecular analyses of MYB and EP300 in 55 patients with myeloproliferative disorders. Blood 2006; 107: 1733–1735, author reply 5.
Ding Y, Harada Y, Imagawa J, Kimura A, Harada H . AML1/RUNX1 point mutation possibly promotes leukemic transformation in myeloproliferative neoplasms. Blood 2009; 114: 5201–5205.
Dey A, Seshasayee D, Noubade R, French DM, Liu J, Chaurushiya MS et al. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science 2012; 337: 1541–1546.
Abdel-Wahab O, Dey A . The ASXL-BAP1 axis: new factors in myelopoiesis, cancer and epigenetics. Leukemia 2013; 27: 10–15.
Ruan HB, Han X, Li MD, Singh JP, Qian K, Azarhoush S et al. O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1alpha stability. Cell Metab 2012; 16: 226–237.
Godfrey AL, Chen E, Pagano F, Ortmann CA, Silber Y, Bellosillo B et al. JAK2V617F homozygosity arises commonly and recurrently in PV and ET, but PV is characterized by expansion of a dominant homozygous subclone. Blood 2012; 120: 2704–2707.
Scott LM, Scott MA, Campbell PJ, Green AR . Progenitors homozygous for the V617F mutation occur in most patients with polycythemia vera, but not essential thrombocythemia. Blood 2006; 108: 2435–2437.
Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu X et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell 2012; 148: 873–885.
Acknowledgements
This work was supported by the NIH Grants K08HL106576 (to STO), K12HL087107 (to STO), P01CA101937 (TJL), and T32HL007088 (to EKE, DACF). This research was also supported by a Sidney Kimmel Scholar Award, Leukemia Research Foundation New Investigator Award, Central Society for Clinical Research Early Career Development Award, Barnes-Jewish Hospital Foundation/Washington University Institute of Clinical and Translational Sciences Pilot Grant, and American Cancer Society Institutional Research Grant (all to STO). This work was supported by the Washington University Institute of Clinical and Translational Sciences Grant UL1TR000448 from the National Center for Advancing Translational Sciences of NIH. Technical support was provided by the Alvin J. Siteman Cancer Center Tissue Procurement and Flow Cytometry Cores, which are supported by NCI Cancer Center Support Grant P30CA91842. We thank D Link and J Xia for assistance with cell sorting and colony assays, K Martin, C Kaiwar and M Fulbright for performing JAK2 and U2AF1 genotyping experiments and J McMichael for assistance with illustrations.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Rights and permissions
About this article
Cite this article
Engle, E., Fisher, D., Miller, C. et al. Clonal evolution revealed by whole genome sequencing in a case of primary myelofibrosis transformed to secondary acute myeloid leukemia. Leukemia 29, 869–876 (2015). https://doi.org/10.1038/leu.2014.289
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2014.289
This article is cited by
-
Phenotypically-defined stages of leukemia arrest predict main driver mutations subgroups, and outcome in acute myeloid leukemia
Blood Cancer Journal (2022)
-
Molecular pathogenesis of the myeloproliferative neoplasms
Journal of Hematology & Oncology (2021)
-
Young versus old age at diagnosis confers distinct genomic profiles in patients with polycythemia vera
Leukemia (2019)
-
Myelofibrosis in 2019: moving beyond JAK2 inhibition
Blood Cancer Journal (2019)
-
Whole-exome sequencing reveals acquisition of mutations leading to the onset of donor cell leukemia after hematopoietic transplantation: a model of leukemogenesis
Leukemia (2018)