Abstract
Inherited retinal degeneration (IRD) are a group of genetically heterogeneous disease of which retinitis pigmentosa (RP) and Leber congenital amaurosis (LCA) are the most common and severe type. In our study we had taken three unrelated South Indian consanguineous IRD families. Homozygosity mapping was done using Affymetrix 250K Nsp1 GeneChip in each of LCA, Cone-Rod dystrophy (CRD) and autosomal recessive RP (arRP) families followed by targeted re-sequencing by next generation sequencing (NGS) on Illumina MiSeq. Known candidate genes ranging from 1–8 in numbers within the homozygous blocks were identified by homozygosity mapping and targeted NGS revealed the causative mutations; RDH12 c.832A>C, ABCA4 c.1462G>T, CDHR1c.1384_1392delCTCCTGGACinsG, in the LCA, CRD and arRP families, respectively. The identified mutations were validated by Sanger sequencing, segregation in the families and their absence in 200 control chromosomes. Homozygosity mapping guided targeted NGS, especially when more numbers of known candidate genes within the homozygous blocks are observed is a comprehensive method for mutation identification. Molecular data from a larger retinal degenerative disease cohort would reveal the spectrum and prevalence of mutations and genes in Indian population. Molecular diagnosis also aids in genetic counseling, offering carrier and prenatal testing to family members.
Similar content being viewed by others
Introduction
Inherited retinal degenerations (IRD) have a prevalence of about 1/3500 in the general population.1 Till date there are about 232 genes that have been identified to be causative of different inherited retinal diseases [RetNet, the Retinal Information Network], and involved in various functions such as phototransduction pathway, the visual cycle, photoreceptor structure, photoreceptor development, transcription factors, cell adhesion, cellular metabolism, protein folding and subunit assembly.2 IRD presents with locus heterogeneity, where mutations in different genes cause the same disease phenotype and allelic heterogeneity as well, where different mutations in the same gene presents different types of retinal disease. Also, IRD follows all Mendelian patterns of inheritance; autosomal dominant, autosomal recessive and X-linked recessive.
The clinical overlap between different IRD is immense. IRD primarily involving the photoreceptors are characterized by progressive loss of rods and then cones (rod-cone dystrophy) or cones followed by rods (cone-rod dystrophy), eventually leading to complete blindness in some. Typical clinical findings in these cases include optic disc pallor, attenuated retinal vessels, pigment dispersal with abnormally diminished or absent a- and b-waves in the electroretinogram (ERG); other associated features include decreased visual acuity, hyperopia or myopia and early onset of posterior subcapsular cataract.
Retinitis pigmentosa (RP) and Leber congenital amaurosis (LCA) are the most common and most severe types of IRD, respectively.3 RP with a prevalence of about 1 in 3000–5000 in the general population, presents with symptoms that include night blindness initially, followed by peripheral visual field defects, tunnel vision and slowly progressing decreased central vision. The age of onset for RP varies between juvenile or early childhood and sixth decade of life.4 In LCA, the affected individuals usually have profound visual loss, nystagmus and non-recordable ERG response within 1 year of life.5 The reported prevalence of LCA is 1:30 000–81 000, and accounts for 5% of all retinal dystrophies.6 There is also a clinical overlap between juvenile RP, diagnosed later to 1 year of age and LCA, where both are categorized as Early Onset Retinal Dystrophy.7 Genes CRB1 and RDH12 are known to cause both LCA and juvenile RP.8 Other IRD genes such as TULP1, SPATA7, KCNJ13, IQCB1 are known to cause LCA and autosomal recessive RP (arRP).9 ABCA4 gene mutations are known to cause various retinal dystrophies like Stargadt’s disease, arRP and autosomal recessive cone-rod dystrophy.10, 11, 12 Owing to this remarkable genetic and phenotypic heterogeneity, accurate molecular diagnosis would aid in the clinical diagnosis, predict the prognosis of the disease, genetic counseling, carrier and prenatal testing.
Homozygosity mapping exploits the fact that stretches of markers along with the disease allele could be homozygous and identical by descent in both consanguineous and non-consanguineous families with autosomal recessive diseases, and hence has been widely used for mapping disease loci/genes.13, 14 Next generation sequencing (NGS) technology uses massively parallel sequencing of candidate genes and recently has been widely used for molecular diagnosis of IRD with a success of mutation identification in about 55–80% of the cases.15 In our earlier report we had screened eleven LCA and one arRP families; molecular analysis was done by homozygosity mapping followed by direct sequencing of candidate gene(s) present within the homozygous blocks. Causative mutations were identified in ten LCA and one arRP families.16 The present study is in continuation of our previous work and here we have performed homozygosity mapping with Affymetrix 250K Nsp1 HMA GeneChip followed by targeted re-sequencing on Illumina Miseq NGS platform in three IRD families. As the number of candidate genes within the homozygous blocks ranged from one to eight, the combined approach was used and was found to be cost-effective and time saving.
Materials and methods
Subjects
Three IRD families; each of LCA, CRD and arRP were studied. Complete ophthalmic examination that included ERG, fundus photograph, fundus auto fluorescence was done in all the seven affected individuals from 3 families. Optical coherence tomography (OCT) was done in three of the affected individuals. OCT was not documented in young/un co-operative children. After obtaining written informed consent, blood samples were collected from all the affected individuals, unaffected siblings and parents. The study was approved by the Institutional Review Board and ethics committee, and all the procedures were in accordance with institutional guidelines and the Declaration of Helsinki.
Homozygosity mapping
Genomic DNA was extracted using Nucleospin Blood XL kit (Macherey-Nagel, GmbH, Düren, Germany). The three IRD families (Fam-01, -02 and -03) were genotyped using 250K NspI Human Mapping Array GeneChip (Affymetrix, Santa Clara, CA, USA) as per the manufacturer’s recommended protocol.
Genotyping was done on one or more affected and at least one unaffected member from each family. The homozygous regions were analyzed using Genotyping Console v4.0 (Affymetrix, Santa Clara, CA, USA) and loss of heterozygosity status was compared between the affected and unaffected. The internal quality control check and loss of heterozygosity scoring was based on our previous report.16 All the homozygous blocks were noted and the known candidate genes within the blocks ranged from 1 to 8. As more than one known candidate gene was shortlisted, we proceeded with targeted re-sequencing using NGS for rapid and cost-effective identification of the causative gene/mutation.
Target region capture and NGS
The targeted sequencing was done for 184 genes associated with eye disorders including retinal diseases at the Strand Centre for Genomics & Personalized Medicine, Bengaluru, India, on Illumina (San Diego, CA, USA) MiSeq platform.
NGS–library preparation and sequencing
We used the Nextera DNA library preparation protocol (Illumina) to convert input genomic DNA (gDNA) into adapter-tagged indexed libraries, which was essentially performed as previously described.17 The quality of tagged and amplified sample libraries was checked by using the BioAnalyzer (Agilent, Santa Clara, CA, USA). The pooled library was loaded and sequenced on the MiSeq platform (Illumina), according to the manufacturer’s instructions.
NGS–data analysis and interpretation
The trimmed FASTQ files were generated using MiSeq Reporter (Illumina). The reads were aligned against the whole genome build: hg19 using Strand NGS v2.1.6 (http://www.strandngs.com/). The data analysis and interpretation were performed by using Strand NGS v2.1.6 and StrandOmics v3.0 (http://www.strandls.com/strandomics/) as previously described.17
Validation of the identified mutations
Validation of the identified mutations was done by PCR and direct Sanger sequencing (ABI 3500 Genetic Analyser (Applied Biosystems, Foster City, CA, USA)) of the specific exons. Further, segregation within the family and screening of 100 healthy control samples was also done to confirm the pathogenicity.
Bioinformatics analyses
The possible effect of missense mutation on structure and function of the protein was anlaysed by PolyPhen-2,18 SIFT,19 Mutation Taster,20 Mutation Accessor,21 PMUT22 and MutPred.23
The identified three novel variations were checked in Human Genome Mutation Database, dbSNP, ClinVar, 1000 Genomes database, Exome Variant Server and the Exome Aggregation Consortium. Also the conservation of the amino acid residue across 46 vertebrates was checked in UCSC database.
Results
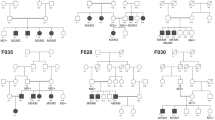
Three consanguineous families; Fam 01—LCA, Fam 02—CRD and Fam 03—arRP, respectively were studied. Totally five affected individuals; two each from Fam 01 and Fam 02 and one from Fam 03, respectively were genotyped using Affymetrix 250K Nsp1 HMA GeneChip. Homozygosity mapping revealed one, eight and four known LCA, arRP and CRD and arRP candidate genes within the homozygous blocks for the families 01, 02 and 03, respectively. Table 1 lists the size of the homozygous blocks and known candidate genes within, for the three families. NGS analysis identified three novel mutations, RDH12 c.832 A>C p.(Ser278Arg), ABCA4 c.1462G>T p.(Glu488Ter) and CDHR1 c.1384_1392delCTCCTGGACinsG p.(Leu462AspfsTer1) in the LCA (Fam 01), CRD (Fam 02) and arRP (Fam 03) families, respectively. Table 2 shows the pathogenic mutations identified in the three families. The mutations segregated in the families and were absent in the 200 control chromosomes screened (Figures 1a–c). Table 3 shows the predicted probable effect of missense mutation in RDH12 using insilico tools. The identified three novel variations are not reported in Human Genome Mutation Database, dbSNP, ClinVar, 1000 Genomes database, Exome Variant Server database and the Exome Aggregation Consortium. The serine residue at 278 position of RDH12 gene was observed to be conserved across vertebrates (UCSC genome browser) except in chicken, zebrafish and lamprey, where it is substituted by a similar hydroxyl group, threonine.
Segregation analysis. (a) Fam 01 LCA RDH12 c.832A>C p.(Ser278Arg), (b) Fam 02 CRD ABCA4 c.1462G>T p.(Glu488Ter), (c) Fam 03 arRP CDHR1 c.1384_1392delCTCCTGGACinsG p.(Leu462AspfsTer1). The arrow indicates the index case. The filled in circles and squares are affected females and males respectively. [M]; [M] – affected with homozygous mutation, [M]; [=] – carries for any given mutation and [=]; [=] – wild type. Lines above the individual indicate availability of genotype. A full color version of this figure is available at the Journal of Human Genetics journal online.
The identified mutations were in the candidate genes present in the homozygous blocks for Fam 02 and Fam 03. For Fam 01 the homozygous block encompassing RDH12 was present in the two affected individuals (the index case and the affected mother), and was 23.9Mb in size and also in the unaffected maternal uncle of about 1.8Mb (encompassing the RDH12 gene). Hence, we did not screen RDH12 gene in this family and screened the other candidate gene, IQCB1. We did not identify any mutation in IQCB1. Following the NGS results and to validate our homozygosity mapping data we genotyped by direct sequencing the entire coding region, and the regions covering the SNPs in the intronic and 5′UTR of RDH12 that are probed in the Affymetrix 250K Nsp1 GeneChip. We genotyped the two affected; the index case and his mother, three unaffected; the index cases’ father, maternal uncle and maternal grandmother. All members were homozygous for all the SNPs in the coding region and for the five SNPs that represent the RDH12 gene in the GeneChip except for the unaffected father who was heterozygous for the SNP at 5′UTR.
Variants of unknown significance identified from NGS
We had also identified variations of unknown significance while performing the NGS analysis.Validation and segeregation for the same was performed in two of the three families, (Fam 01 and Fam 03) to know if there was any modifier effect (Table 4). For Fam 02, few variations of unknown significance in ZNF513, ROM1, OPA1, HMCN1 was observed but validation and segregation analysis was not performed for these.
Phenotype features
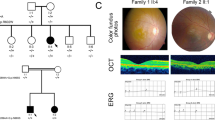
Fam 01. In this LCA family, the affected proband and his mother had novel missense mutation c.832A>C p.(Ser278Arg) in RDH12. The proband had history of profound visual loss with night blindness and nystagmus since birth. The fundus of the mother (40 years) also showed pale disc and attenuated vessels with well-demarcated area of macular atrophy, extensive widespread bony spicule pigment were seen in the periphery as well as the macula (Figure 2a). OCT of the same patient (Figure 2b) also revealed gross foveal thining at the macula in both eyes with the pigments in macular area showing extensive shadowing. The proband (20 years) also showed pale disc, attenuated vessels with extensive peripheral as well as macular pigment (Figure 2c). The macula had a typical ‘petal like macular coloboma’ with sloping sides and central excavation and pigments, which was also seen on OCT (Figure 2d). The mother and proband had an extinguished photopic and scotopic response in ERG.
Fundus photographs. (a) A 40 year old female with c.832A>C p.(Ser278Arg) mutation in RDH12 (Fam 01,proband’s mother) showed pale disc, attenuated vessels, well defined macular atrophy wide spread bony spicule pigment seen in the periphery as well as macula. (b) OCT (Fam 01 proband’s mother) in the left eye showed shadowing because of macular pigment. (c) A 20 year old male with c.832A>C p.(Ser278Arg) mutation in RDH12 (Fam 01, proband) showed pale disc, attenuated vessels, prominent petal like macular coloboma with sloping sides and plenty of peripheral pigmentation. (d) OCT (Fam 01, proband) in the right eye showed extensive macular coloboma. A full color version of this figure is available at the Journal of Human Genetics journal online.
Fam 02. In this CRD family, all the five affected individuals had novel ABCA4 nonsense mutation c.1462G>T p.(Glu488Ter). All five patients had a profound visual loss with difficulty in night vision. The proband (15 years) had well-delineated RPE atrophy and pigment dispersal at the posterior pole (Figure 3a). The proband’s mother (40 years) had an extensive RPE atrophy and widespread pigment dispersal (Figure 3b). ERG of the proband and mother (Figures 3c and d) showed progressive decrease in the amplitudes of photopic and scotopic response. Proband’s grandfather (67 years) and maternal uncle (41 years) also had a picture of advanced disease with extinguished ERG. Since the photopic responses were reduced first (as seen in the proband) and the scotopic responses were found to be extinguished in older members of the family, a diagnosis of progressive autosomal recessive cone-rod dystrophy was made.
Fundus photograph and ERG Trace. (a) A 15 year old female with c.1462G>T p.(Glu488Ter) mutation in ABCA4 (Fam 02, Fundus montage of the left eye in proband) showed well delineated RPE atrophy and pigment dispersal at the posterior pole. (b) A 40 year old female with c.1462G>T p.(Glu488Ter) mutation in ABCA4 (Fam 02, Fundus montage of the right eye in proband’s mother) showed extensive RPE atrophy and widespread pigment dispersal. (c, d) ERG pictures of the proband 15 years (c) and proband’s mother 40 years (d) showed progressive decrease in the amplitudes of photopic and scotopic response. A full color version of this figure is available at the Journal of Human Genetics journal online.
Fam 03. A novel indel was identifed in CDHR1 c.1384_1392delCTCCTGGACinsG in this case of arRP. The proband (27 years) had history of night blindness since 8 years with myopic refraction and fundus picture showed pale disc, attenuated vessels with atrophic macula and yellowish spots in the periphery (Figure 4). ERG findings revealed non-recordable scotopic and photopic responses.
Fundus photograph. A 27-year-old female with c.1384_1392delCTCCTGGACinsG p.(Leu462AspfsTer1) in CDHR1 (Fam 03) showed disc palor, arteriolar attenuation, atrophic macula with yellowish spots in the periphery. A full color version of this figure is available at the Journal of Human Genetics journal online.
Discussion
In this study on three consanguineous recessive families diagnosed with inherited retinal disease, the disease causing mutation was identified in all. Homozygosity mapping revelaved homozygous blocks of size 1–40 Mb, and the number of known candidate genes within homozygous blocks ranged from 1 to 8. Following homozygosity mapping, targeted re-sequencing of retinal disease genes by NGS was performed in the proband of all the families for rapid identification of the causative gene/mutation. The causative mutation identified was present in the gene within the homozygous blocks for Fam 02 and 03.
In the Fam 01, totally 32 homozygous blocks were shared between the two affected, with the largest block being 23.9 Mb and the smallest 0.2 Mb. An 8Mb block shared exclusively by the two affected, harbouring IQCB1 gene was the only known LCA candidate gene. As screening of IQCB1 did not identify any mutation, we proceeded with targeted NGS. Identification of novel mutation in RDH12 in the proband and validation followed by segregation in the family confirming the NGS results lead us to re-analyze the homozygosity mapping data. The re-analysis explained the smaller homozygous block of 1.8 Mb encompassing the RDH12 gene in the unaffected maternal uncle. On an average 11% of genome of individuals with recessive disease and having first cousins as parents is homozygous with at least 20 homozygous blocks measuring >3 cM. Also the number and runs of homozygosity is larger in consanguineous mating.24, 25 Hildebrandt et al. have demonstrated that homozygosity mapping with chip density of 250 K can identify recessive disease genes in ~2 Mb region even in an outbred population.26 In this case (Fam 01), a higher density chip would have revealed closer regions of recombination that would have helped us picking RDH12 for screening. The identified novel missense mutation in RDH12 c.832A>C p.(Ser278Arg) segregated with the disease in the family was absent in 200 control chromosomes, the serine residue at position 278 is conserved across most of the vetebrates and also five of the six in silico analyzes predict the mutation to be damaging. However, in vitro functional analysis of the catalytic activity of the identified missense mutation would further confirm the pathogenicity of the mutation.27 Phenotypically both the proband and mother had typical petal coloboma, a characteristic feature of RDH12 mutation28 and thus the novel missense mutation is defined as likely pathogenic.
In Fam 02 with novel nonsense mutation in ABCA4 c.1462G>T p.(Glu488Ter), the younger members of this family showed preserved scotopic responses but the older members had a progressive decrease in the photopic, as well as the scotopic response. Similar progression has been described in ABCA4 mutations.29 The phenotypic variability within the family could be because of progression with age11 and modifying effects, either environmental or genetic or both.30
In Fam 03, we had identified a novel indel in CDHR1 c.1384_1392delCTCCTGGACinsG. Mutations in CDHR1 gene has been reported in autosomal recessive cone-rod dystrophy or retinal dystrophy and till date seven families have been decribed.31 Here, for the first time we report CDHR1 mutation in a case of arRP from India. The fundus showed attenuated vessels with atrophic macula and yellowish spots in the periphery similar to earlier report of retinal dystrophy with CDHR1 mutation.31
As mentioned variations of unknown significance were observed in these families, some of which have been validated and checked for segregation. However, the phenotype or the severity did not specifically show any distinct feature that could be attributed to the modifier effect of the variations of unknown significance, and hence their contribution or role is not clear.
In summary, in this study we had performed homozygosity mapping in three IRD families using Affymetrix 250K NspI GeneChip and since the number of known candidate genes in the homozygous blocks were more, it was followed by targeted NGS. We have identified three novel mutations, one each in RDH12, ABCA4, CDHR1 gene by this combined approach. The molecular diagnosis in these families helps in appropriate counselling and in prognosis prediction.
References
Shanks, M. E., Downes, S. M., Copley, R. R., Lise, S., Broxholme, J., Hudspith, K. A. et al. Next-generation sequencing (NGS) as a diagnostic tool for retinal degeneration reveals a much higher detection rate in early-onset disease. Eur. J. Hum. Genet. 21, 274–280 (2013).
Jin, X., Qu, L. H., Meng, X. H., Xu, H. W. & Yin, Z. Q. Detecting genetic variations in hereditary retinal dystrophies with next-generation sequencing technology. Mol. Vis. 20, 553–560 (2014).
Rivolta, C., Sharon, D., DeAngelis, M. M. & Dryja, T. P. Retinitis pigmentosa and allied diseases: numerous diseases, genes, and inheritance patterns. Hum. Mol. Genet. 11, 1219–1227 (2002).
Fahim, A. T., Daiger, S. P. & Weleber, R. G. in Retinitis Pigmentosa Overview (eds Pagon, R. A., Adam, M. P., Ardinger, H. H., Wallace, S. E., Amemiya, A., Bean, L. J. H. et al.) (Seattle, WA, USA, 1993).
Dharmaraj, S. R., Silva, E. R., Pina, A. L., Li, Y. Y., Yang, J. M., Carter, C. R. et al. Mutational analysis and clinical correlation in Leber congenital amaurosis. Ophthalmic Genet. 21, 135–150 (2000).
Chacon-Camacho, O. F. & Zenteno, J. C. Review and update on the molecular basis of Leber congenital amaurosis. World J. Clin. Cases 3, 112–124 (2015).
Gu, S. M., Thompson, D. A., Srikumari, C. R., Lorenz, B., Finckh, U., Nicoletti, A. et al. Mutations in RPE65 cause autosomal recessive childhood-onset severe retinal dystrophy. Nat. Genet. 17, 194–197 (1997).
den Hollander, A. I., Lopez, I., Yzer, S., Zonneveld, M. N., Janssen, I. M., Strom, T. M. et al. Identification of novel mutations in patients with Leber congenital amaurosis and juvenile RP by genome-wide homozygosity mapping with SNP microarrays. Invest. Ophthalmol. Vis. Sci. 48, 5690–5698 (2007).
den Hollander, A. I., Roepman, R., Koenekoop, R. K. & Cremers, F. P. Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog. Retin. Eye Res. 27, 391–419 (2008).
Koenekoop, R. K. The gene for Stargardt disease, ABCA4, is a major retinal gene: a mini-review. Ophthalmic Genet. 24, 75–80 (2003).
Mullins, R. F., Kuehn, M. H., Radu, R. A., Enriquez, G. S., East, J. S., Schindler, E. I. et al. Autosomal recessive retinitis pigmentosa due to ABCA4 mutations: clinical, pathologic, and molecular characterization. Invest. Ophthalmol. Vis. Sci. 53, 1883–1894 (2012).
Valverde, D., Riveiro-Alvarez, R., Aguirre-Lamban, J., Baiget, M., Carballo, M., Antinolo, G. et al. Spectrum of the ABCA4 gene mutations implicated in severe retinopathies in Spanish patients. Invest. Ophthalmol. Vis. Sci. 48, 985–990 (2007).
Kannabiran, C., Singh, H., Sahini, N., Jalali, S. & Mohan, G. Mutations in TULP1, NR2E3, and MFRP genes in Indian families with autosomal recessive retinitis pigmentosa. Mol. Vis. 18, 1165–1174 (2012).
Ramprasad, V. L., Soumittra, N., Nancarrow, D., Sen, P., McKibbin, M., Williams, G. A. et al. Identification of a novel splice-site mutation in the Lebercilin (LCA5) gene causing Leber congenital amaurosis. Mol. Vis. 14, 481–486 (2008).
Glockle, N., Kohl, S., Mohr, J., Scheurenbrand, T., Sprecher, A., Weisschuh, N. et al. Panel-based next generation sequencing as a reliable and efficient technique to detect mutations in unselected patients with retinal dystrophies. Eur. J. Hum. Genet. 22, 99–104 (2014).
Srilekha, S., Arokiasamy, T., Srikrupa, N. N., Umashankar, V., Meenakshi, S., Sen, P. et al. Homozygosity Mapping in Leber Congenital Amaurosis and Autosomal Recessive Retinitis Pigmentosa in South Indian Families. PLoS ONE 10, e0131679 (2015).
Mannan, A. U., Singh, J., Lakshmikeshava, R., Thota, N., Singh, S., Sowmya, T. S. et al. Detection of high frequency of mutations in a breast andor ovarian cancer cohort: implications of embracing a multi-gene panel in molecular diagnosis in India. J. Hum. Genet. 61, 515–522 (2016).
Adzhubei, I. A., Schmidt, S., Peshkin, L., Ramensky, V. E., Gerasimova, A., Bork, P. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Kumar, P., Henikoff, S. & Ng, P. C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4, 1073–1081 (2009).
Schwarz, J. M., Cooper, D. N., Schuelke, M. & Seelow, D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods 11, 361–362 (2014).
Reva, B., Antipin, Y. & Sander, C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 39, e118 (2011).
Ferrer-Costa, C., Gelpi, J. L., Zamakola, L., Parraga, I., de la Cruz, X. & Orozco, M. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics 21, 3176–3178 (2005).
Li, B., Krishnan, V. G., Mort, M. E., Xin, F., Kamati, K. K., Cooper, D. N. et al. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics 25, 2744–2750 (2009).
Woods, C. G., Cox, J., Springell, K., Hampshire, D. J., Mohamed, M. D., McKibbin, M. et al. Quantification of homozygosity in consanguineous individuals with autosomal recessive disease. Am. J. Hum. Genet. 78, 889–896 (2006).
Kirin, M., McQuillan, R., Franklin, C. S., Campbell, H., McKeigue, P. M. & Wilson, J. F. Genomic runs of homozygosity record population history and consanguinity. PLoS ONE 5, e13996 (2010).
Hildebrandt, F., Heeringa, S. F., Ruschendorf, F., Attanasio, M., Nurnberg, G., Becker, C. et al. A systematic approach to mapping recessive disease genes in individuals from outbred populations. PLoS Genet. 5, e1000353 (2009).
Thompson, D. A., Janecke, A. R., Lange, J., Feathers, K. L., Hubner, C. A., McHenry, C. L. et al. Retinal degeneration associated with RDH12 mutations results from decreased 11-cis retinal synthesis due to disruption of the visual cycle. Hum. Mol. Genet. 14, 3865–3875 (2005).
Valverde, D., Pereiro, I., Vallespin, E., Ayuso, C., Borrego, S. & Baiget, M. Complexity of phenotype-genotype correlations in Spanish patients with RDH12 mutations. Invest. Ophthalmol. Vis. Sci. 50, 1065–1068 (2009).
Maugeri, A., Klevering, B. J., Rohrschneider, K., Blankenagel, A., Brunner, H. G., Deutman, A. F. et al. Mutations in the ABCA4 (ABCR) gene are the major cause of autosomal recessive cone-rod dystrophy. Am. J. Hum. Genet. 67, 960–966 (2000).
Cideciyan, A. V., Swider, M., Aleman, T. S., Tsybovsky, Y., Schwartz, S. B., Windsor, E. A. et al. ABCA4 disease progression and a proposed strategy for gene therapy. Hum. Mol. Genet. 18, 931–941 (2009).
Nikopoulos, K., Avila-Fernandez, A., Corton, M., Lopez-Molina, M. I., Perez-Carro, R., Bontadelli, L. et al. Identification of two novel mutations in CDHR1 in consanguineous Spanish families with autosomal recessive retinal dystrophy. Sci. Rep. 5, 13902 (2015).
Acknowledgements
We thank Indian Council of Medical Research (ICMR), Govt. of India for the grant 54/1/2010-BMS and SRF Fellowship – N0.45/2/2014-HUM-BMS, and all the patients and their family for their kind cooperation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Sundaramurthy, S., Swaminathan, M., Sen, P. et al. Homozygosity mapping guided next generation sequencing to identify the causative genetic variation in inherited retinal degenerative diseases. J Hum Genet 61, 951–958 (2016). https://doi.org/10.1038/jhg.2016.83
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2016.83