Abstract
Tooth agenesis is the most common developmental anomaly of human dentition, occurring most often in the third molar (wisdom tooth). It is affected by genetic variation, so this study aimed to identify susceptibility genes associated with third molar agenesis. Examination of panoramic radiographs and medical history about third molar extraction were used to diagnose third molar agenesis. We then conducted a genome-wide association study of 149 cases with at least one-third molar agenesis and 338 controls from Japan and Korea using the Illumina HumanOmniExpress BeadChip. After rigorous quality-control filtering, approximately 550 000 single-nucleotide polymorphisms (SNPs) were analyzed in association tests with the status. We identified three SNPs showing evidence of association at P<1 × 10−5 and 69 SNPs showing evidence of association at P<1 × 10−4. SNP rs1469622, which maps to an intron of THSD7B (thrombospondin, type I, domain containing 7B) on chromosome 2, showed the strongest association (combined odds ratio=1.88, 95% confidence interval=1.43–2.47, P=7.5 × 10−6). The identified SNPs may be considered candidates for future replication studies in independent samples.
Similar content being viewed by others
Introduction
Tooth agenesis is a common human anomaly, with that of the third molar (wisdom tooth) occurring most often. Tooth agenesis of the mandibular central and lateral permanent incisors has the lowest incidence.1 The incidences of third molar agenesis vary between populations,2 reaching nearly 100% in Mexican Indians and being almost absent in Tasmanians. The number of missing third molars in individuals with any missing third molar (mean, 1.74 per person) is significantly lower in blacks than whites (mean, 2.05).3
Genetic components appear to be strongly involved in tooth agenesis. Variants in several genes including msh homeobox 1 (MSX1),4 paired box 9 (PAX9)5 and axin 2 (AXIN2)6 have been reported as causal factors of non-syndromic hypodontia (missing up to six teeth, excluding the third molars) or oligodontia (missing over six teeth, excluding the third molars). MSX1 is associated with cleft palate,7 whereas AXIN2 is associated with colorectal cancer.6 Although some genes related to early tooth development are associated with tooth agenesis and systemic features,8 the susceptibility genes or loci for third molar agenesis remain unknown. The identification of such genes is likely to contribute to the fields of medicine, dentistry, anthropology, anatomy and phylogenesis.
In this study, we conducted a genome-wide association study (GWAS) of third molar agenesis. We examined 149 cases with at least one-third molar agenesis and 338 control subjects without. We identified three single-nucleotide polymorphisms (SNPs) showing evidence of association at P<1 × 10−5 and 69 SNPs showing evidence of association at P<1 × 10−4. Our results therefore provide informative candidates for further follow-up studies.
Materials and methods
Study participants and diagnosis
A total of 503 individuals were enrolled in the study, including unrelated Japanese individuals from the Tokyo metropolitan area, and unrelated Korean individuals from the Busan area, South Korea. The study was approved by the Institutional Review Board of each institution, and informed consent was obtained from all participating individuals.
Completely calcified crowns are reported to be visible around the age of 13–15 years,9, 10, 11 so we enrolled participants aged older than 16 years. Diagnosis of third molar agenesis was made by examination of a panoramic radiograph12 (Figure 1) taken between 2008 and 2012 at Showa University Dental Hospital, Tokyo, or Pusan National University Dental Hospital, Busan, together with interviews about the medical history of third molar extraction. If we were unable to confirm whether third molar(s) were extracted, the subjects were excluded from the study. The control subjects were defined by confirming the presence of four third molars on a panoramic radiograph.
Subjects with at least one-third molar agenesis included 78 Japanese (17 men, 61 women) and 71 Koreans (39 men, 32 women). Controls without third molar agenesis included 186 Japanese (57 men, 129 women) and 152 Koreans (91 men, 61 women). The mean age of the individuals was 26.3 years (range, 16–57 years) for the Japanese, and 27.9 years (range, 20–40 years) for the Koreans.
DNA collection, genotyping and quality control
Saliva was collected from study participants and DNA was isolated using an Oragene DNA self-collection kit (DNA Genotek, Ottawa, ON, Canada), following the manufacturer’s instructions. Aliquots containing 200 ng total DNA per sample were used. All DNA samples from the Japanese and Korean subjects were analyzed with a HumanOmniExpress BeadChip (Illumina, San Diego, CA, USA) according to the manufacturer’s protocol. Genotype calling of more than 700 000 SNPs was implemented using Illumina BeadStudio software (Illumina). All genotyping assays were performed at Kyushu University (Fukuoka, Japan).
We applied the following quality control (QC) procedures separately to the Japanese and Korean data sets using the PLINK version 1.07 software.13 The outline of our QC filtering is shown in Supplementary Figure S1. We confirmed that the information for the gender of each individual was consistent with the SNP genotyping data on the X chromosome. We then excluded SNPs on sex chromosomes. We excluded subjects with missing genotype rates >2% and SNPs with missing genotype rates >2%. We included SNPs showing minor allele frequencies ⩾3% and excluded those showing significant deviations from the Hardy–Weinberg equilibrium at P<1 × 10−6. The significance threshold of the P-value for excluding SNPs caused by deviation from Hardy–Weinberg equilibrium was determined by analyzing the relationship between the distribution of the P-values and the over- and under-representation of heterozygotes (Supplementary Figure S2).
In terms of the identical-by-descent probability, we detected a pair of subjects with cryptic relatedness whose genomes were shared with each other at the expected levels of sharing between parent and offspring or full siblings. We randomly selected and retained one subject out of the relatives for further analyses. We performed a principal component analysis including the Japanese and Korean data sets together with HapMap phase III samples (2010) using the EIGENSTRAT 3.0 software.14, 15 As a result, we found that all subjects in our data sets were of East Asian origin (Supplementary Figure S3a). A principal component analysis using only East Asian populations showed that some of our Japanese subjects were in non-Japanese clusters: two Chinese, three Korean, and four Japanese–Chinese mixed clusters (Supplementary Figure S3b). We removed these nine subjects from further analyses.
Statistical analysis
We defined three types of phenotypes regarding third molar agenesis: (A) individuals with at least one-third molar agenesis were treated as cases and individuals with four third molars as controls (Japanese cases 78, controls 186; Korean cases 71, controls 152), (B) individuals with and without maxillary third molar agenesis were treated as cases and controls, respectively (Japanese cases 62, controls 202; Korean cases 63, controls 160), (C) individuals with and without mandibular third molar agenesis were treated as cases and controls, respectively (Japanese cases 40, controls 224; Korean cases 40, controls 183).
We initially analyzed the Japanese and Korean data sets separately, and then combined the results from the two data sets by means of meta-analysis16 using the PLINK software.13 The initial association analyses were performed using a 2 × 2 contingency table based on allele frequency. For all filtered SNPs, the P-values of association were assessed by Fisher’s exact test, and odds ratios (ORs) and 95% confidence intervals (95% CIs) were calculated. The quantile–quantile plot and genomic inflation factor (λ) were used to assess the presence of systematic bias of the observed test statistics based on the potential population structure. Second, we conducted random-effects model meta-analyses17 by combining the results from the Japanese and Korean data sets. We evaluated the between-study heterogeneity in terms of Cochran’s Q test18 and I2 metrics.19 The meta-analyses and evaluation of heterogeneity were rerun using the STATA version 11.0 software (Stata, College Station, TX, USA) for confirmation. A total of 532 105 SNPs, common to the filtered Japanese and Korean data sets, were used in the meta-analysis.
According to the statistical power evaluation (Supplementary Figure S4), our GWAS was found to have substantial power (>0.5) to detect genetic variants with risk allele frequencies ranging from 0.2 to 0.75 and relatively large effect sizes (OR=2.0) at the significant threshold of P<10−4. At the same time, the result of the power estimation shows that it is difficult to identify genetic variants at the stringent significance level (P<5 × 10−8) unless their effects are very strong (OR⩾2.5). Therefore, we defined P<10−4 as a threshold for reporting suggestive associations for future replication studies.
Results
Association studies in individual data sets
After QC filtering, 547 727 SNPs and 545 945 SNPs were available within the Japanese and Korean data sets, respectively. The average genotype call rates were >99.9% in both data sets. The quantile–quantile plots for (A) at least one-third molar agenesis, (B) maxillary third molar agenesis and (C) mandibular third molar agenesis are shown in Supplementary Figure S5. Genomic inflation factors obtained for all analyses were <1.03, indicating that the observed test statistics were not inflated by a systematic bias such as population stratification. Forty-nine and 34 SNPs showed significant associations at P<1 × 10−4 in the Japanese and Korean data sets, respectively, for at least one-third molar agenesis; 45 and 41 SNPs showed significant associations at P<1 × 10−4 in the Japanese and Korean data sets, respectively, for maxillary third molar agenesis; 36 and 24 SNPs showed significant associations at P<1 × 10−4 in the Japanese and Korean data sets, respectively, for mandibular third molar agenesis.
Meta-analysis
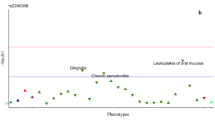
To improve the statistical power and detect consistent association signals, we combined the Japanese and Korean data sets using meta-analysis. The results of random-effects model meta-analysis for the three types of third molar agenesis phenotypes are shown in Figure 2. We detected 21, 26 and 30 SNPs showing significant associations at P<1 × 10−5 in the meta-analyses for at least one, maxillary and mandibular third molar agenesis, respectively (Supplementary Table S1). Of these, three SNPs showed significant associations at P<1 × 10−5 (Table 1). SNP rs1469622 showed the strongest association with at least one-third molar agenesis (combined OR=1.88, 95% CI=1.43–2.47, P=7.5 × 10−6). This SNP maps to the intron of the thrombospondin, type I, domain containing 7B gene (THSD7B) on chromosome 2. SNPs rs938036 and rs906628 showed significant associations with mandibular third agenesis at P=7.8 × 10−6 and 8.3 × 10−6, respectively.
Manhattan plot of the random-effects model meta-analysis of combined genome-wide association study (GWAS) results from Japanese and Korean data sets. (a) Manhattan plot of the combined Japanese and Korean data set meta-analysis between individuals with at least one-third molar agenesis and those without. (b) Manhattan plot of the combined Japanese and Korean data set meta-analysis between individuals with and without maxillary third molar agenesis. (c) Manhattan plot of the combined Japanese and Korean data set meta-analysis between individuals with and without mandibular third molar agenesis.
Discussion
Our GWAS identified three genomic regions showing evidence of association (P<1 × 10−5) with third molar agenesis; the strongest association (combined OR=1.88, 95% CI=1.43–2.47, P=7.5 × 10−6) was found with SNP rs1469622, which maps to the intron of THSD7B. SNPs in this gene were shown in a recent GWAS to be associated with bipolar disorder (rs718632),20 pancreatic cancer in a Japanese population (rs1427593)21 and alcohol dependence (rs1869324).22 However, the biological function of THSD7B is unknown.23 In this study, SNPs rs938036 and rs906628 both showed evidence of association with third molar agenesis at P<1 × 10−5. The nearest genes to rs938036 and rs906628 are LOC100420828 and LOC100507548, respectively; however, the biological functions of these genes are again unknown.
Of the SNPs showing evidence of association at P<1 × 10−4, meta-analysis identified four coincident SNPs in at least one (A) and maxillary (B) third molar agenesis (rs1469622, rs6459434, rs12369725, rs1075010), one coincident SNP in at least one (A) and mandibular (C) third molar agenesis (rs10834449), and one coincident SNP in maxillary (B) and mandibular (C) third molar agenesis (rs1339800). SNP rs12369725, which maps to the intron of the glutamate receptor, ionotropic, N-methyl D-aspartate 2B gene (GRIN2B) on chromosome 12p12 also showed an association (OR=2.44, 95% CI=1.50–3.98 in Japanese subjects, OR=1.86, 95% CI=1.16–2.97 in Korean subjects, OR=2.12, 95% CI=1.51–2.97, P=1.4 × 10−5 in the combined analyses). Previously, SNP rs2284411 in GRIN2B showed an association in attention-deficit hyperactivity disorder families.24 Moreover, recent trio-based exome sequencing suggested that de novo mutations in GRIN2B may contribute substantially to the genetic risk for autism spectrum disorders,25 whereas a GWAS showed that rs220549 in GRIN2B had an association with neuroticism.26
SNP rs10834449, which maps to the intron of the leucine zipper protein 2 gene (LUZP2) on chromosome 11p14.3, also showed an association with third molar agenesis (combined OR=1.84 and 2.14, 95% CI=1.39–2.43 and 1.49–3.07, P=2.2 and 4.3 × 10−5). This gene is deleted in some patients with Wilms tumor, aniridia, genitourinary anomalies-mental retardation (WAGR) syndrome.27 Moreover, SNP rs3734652, which maps to the 3′-untranslated region of ZBTB24 on chromosome 6, showed an association with third molar agenesis (combined OR=2.03, 95% CI=1.44–2.87, P=6.2 × 10−5). ZBTB24 was recently shown to be responsible for immunodeficiency, centromeric instability and facial anomalies (ICF), type 2.28 Finally, SNP rs492761, which maps 3′ downstream of THYN1 on chromosome 11, showed an association with third molar agenesis (combined OR=1.90, 95% CI=1.40–2.59, P=3.9 × 10−5). THYN1 is a candidate gene for developmental delay/mental retardation.29
Genetic factors are strongly involved in non-syndromic tooth agenesis, and several genes have been reported to carry mutations or variants including MSX1 (second premolars and third molars),4 PAX9 (oligodontia)5 and AXIN2 (oligodontia). Mutations in MSX1 cause tooth agenesis of various types of teeth, including second premolars, third molars and incisors, and has been associated with hypodontia of mainly second premolars and third molars in families.4, 30, 31 Conversely, MSX1 has been excluded from other forms of hypodontia involving both second premolars, as the cause for the more common cases of tooth agenesis where only one or two teeth are missing is not explained by MSX1 mutations.32
Third molar agenesis is more commonly found in the maxilla than in the mandible,8, 33, 34, 35, 36, 37 or in both jaws.38 In patients with severe hypodontia, patterns of tooth agenesis in the maxilla and mandible are independent.39 The mechanism underlying the difference in third molar agenesis between the maxilla and mandible is not clear.34, 38 Differential expression of several genes indicates a genetic difference between tooth specification and the molecular pathways that control tooth formation in the maxilla and mandible.40 The maxillary and mandibular ectomesenchyme responds differently to ectodermal signaling for certain genetic pathways;41 therefore, we defined three phenotypes, and analyzed them separately.
Approximately 20 more oligodontia or hypodontia-causing PAX9 mutations have been reported, most of which are located in the paired domain. The mutations range from missense mutations to premature terminations and deletion of the entire gene. They are all heterozygous, indicating that the mechanism of haploinsufficiency has a role in tooth agenesis.42, 43 Mutations in AXIN2 cause tooth agenesis that predominantly affects permanent teeth,8 including permanent molars, lower incisors and upper lateral incisors in one large family and in a sporadic case.6 According to the results of our present meta-analysis, these genes did not show significant associations at P<1 × 10−4 (Supplementary Table S2).
In conclusion, we have conducted the first GWAS to identify susceptibility genes underlying third molar agenesis diagnosed by examination of a panoramic radiograph. As a result, we identified three SNPs showing association signals with third molar agenesis at P<1 × 10−5 located in three independent loci. We did not find any SNPs showing significant associations at P<1 × 10−4 among genes previously reported to be associated with non-syndromic permanent tooth agenesis, suggesting that there might be differences in developmental mechanics between third molar agenesis and other permanent tooth agenesis. Although our GWAS used samples from East Asian populations, the prevalence of third molar agenesis is known to vary across populations.2, 3, 44, 45 Moreover, this study is only a first-stage investigation; therefore, SNPs with significant associations should be considered candidates for future studies to investigate the potential mechanisms underlying racial or ethnic differences. Nevertheless, these findings advance our understanding of the genetic basis of third molar agenesis.
References
Silva, M. R. Radiographic assessment of congenitally missing teeth in orthodontic patients. Int. J. Paediatr. Dent. 13, 112–116 (2003).
Rozkovcová, E., Marková, M. & Dolejsí, J. Studies on agenesis of third molars amongst populations of different origin. Sb. Lek. 100, 71–84 (1999).
Harris, E. F. & Clark, L. L. Hypodontia: an epidemiologic study of American black and white people. Am. J. Orthod. Dentofacial Orthop. 134, 761–767 (2008).
Vastardis, H., Karimbux, N., Guthua, S. W., Seidman, J. G. & Seidman, C. E. A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat. Genet. 13, 417–421 (1996).
Stockton, D. W., Das, P., Goldenberg, M., D'Souza, R. N. & Patel, P. I. Mutation of PAX9 is associated with oligodontia. Nat. Genet. 24, 18–19 (2000).
Lammi, L., Arte, S., Somer, M., Jarvinen, H., Lahermo, P., Thesleff, I. et al. Mutations in AXIN2 cause familial tooth agenesis and predispose to colorectal cancer. Am. J. Hum. Genet. 74, 1043–1050 (2004).
van den Boogaard, M. J., Dorland, M., Beemer, F. A. & van Amstel, H. K. MSX1 mutation is associated with orofacial clefting and tooth agenesis in humans. Nat. Genet. 24, 342–343 (2000).
Bailleul-Forestier, I., Molla, M., Verloes, A. & Berdal, A. The genetic basis of inherited anomalies of the teeth. Part 1: clinical and molecular aspects of non-syndromic dental disorders. Eur. J. Med. Genet. 51, 273–291 (2008).
Karadayi, B., Kaya, A., Kolusayın, M. O., Karadayi, S., Afsin, H. & Ozaslan, A. Radiological age estimation: based on third molar mineralization and eruption in Turkish children and young adults. Int. J. Legal Med. 126, 933–942 (2012).
Harris, E. F. Mineralization of the mandibular third molar: a study of American blacks and whites. Am. J. Phys. Anthropol. 132, 98–109 (2007).
Bolaños, M. V., Moussa, H., Manrique, M. C. & Bolaños, M. J. Radiographic evaluation of third molar development in Spanish children and young people. Forensic Sci. Int. 133, 212–219 (2003).
Baba-Kawano, S., Toyoshima, Y., Regalado, L., Sa'do, B. & Nakasima, A. Relationship between congenitally missing lower third molars and late formation of tooth germs. Angle Orthod. 72, 112–117 (2002).
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Patterson, N., Price, A. L. & Reich, D. Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006).
Price, A. L., Patterson, N. J., Plenge, R. M., Weinblatt, M. E., Shadick, N. A. & Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Nakaoka, H. & Inoue, I. Meta-analysis of genetic association studies: methodologies, between-study heterogeneity and winner’s curse. J. Hum. Genet. 54, 615–623 (2009).
DerSimonian, R. & Laird, N. Meta-analysis in clinical trials. Control Clin. Trials 7, 177–188 (1986).
Cochran, W. G. The combination of estimates from different experiments. Biometrics 10, 101–129 (1954).
Higgins, J. P., Thompson, S. G., Deeks, J. J. & Altman, D. G. Measuring inconsistency in meta-analyses. BMJ 327, 557–560 (2003).
Hattori, E., Toyota, T., Ishitsuka, Y., Iwayama, Y., Yamada, K., Ujike, H. et al. Preliminary genome-wide association study of bipolar disorder in the Japanese population. Am. J. Med. Genet. B 150B, 1110–1117 (2009).
Low, S. K., Kuchiba, A., Zembutsu, H., Saito, A., Takahashi, A., Kubo, M. et al. Genome-wide association study of pancreatic cancer in Japanese population. PLoS One 5, e11824 (2010).
Wang, K. S., Liu, X., Zhang, Q., Pan, Y., Aragam, N. & Zeng, M A meta-analysis of two genome-wide association studies identifies 3 new loci for alcohol dependence. J. Psychiatr. Res. 45, 1419–1425 (2011).
Gobeil, S., Zhu, X., Doillon, C. J. & Green, M. R. A genome-wide shRNA screen identifies GAS1 as a novel melanoma metastasis suppressor gene. Genes Dev. 22, 2932e40 (2008).
Dorval, K. M., Wigg, K. G., Crosbie, J., Tannock, R., Kennedy, J. L., Ickowicz, A. et al. Association of the glutamate receptor subunit gene GRIN2B with attention-deficit/hyperactivity disorder. Genes Brain Behav. 6, 444–452 (2007).
O'Roak, B. J., Deriziotis, P., Lee, C., Vives, L., Schwartz, J. J., Girirajan, S. et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat. Genet. 43, 585–589 (2011).
Aragam, N., Wang, K. S., Anderson, J. L. & Liu, X. TMPRSS9 and GRIN2B are associated with neuroticism: a genome-wide association study in a European sample. J. Mol. Neurosci. 50, 250–256 (2013).
Wu, M., Michaud, E. J. & Johnson, D. K. Cloning, functional study and comparative mapping of Luzp2 to mouse chromosome 7 and human chromosome 11p13–11p14. Mamm. Genome 14, 323–334 (2003).
de Greef, J. C., Wang, J., Balog, J., den Dunnen, J. T., Frants, R. R., Straasheijm, K. R. et al. Mutations in ZBTB24 are associated with immunodeficiency, centromeric instability, and facial anomalies syndrome type 2. Am. J. Hum. Genet. 88, 796–804 (2011).
Taoyun, J., Ye, W., Huifang, W., Jingmin, W. & Yuwu, J. Diagnosis and fine mapping of a deletion in distal 11q in two Chinese patients with developmental delay. J. Hum. Genet. 55, 486–489 (2010).
Nieminen, P., Arte, S., Pirinen, S., Peltonen, L. & Thesleff, I. Gene defect in hypodontia: exclusion of MSX1 and MSX2 as candidate genes. Hum. Genet. 96, 305–308 (1995).
Mostowska, A., Biedziak, B. & Jagodzinski, P. P. Novel MSX1 mutation in a family with autosomal-dominant hypodontia of second premolars and third molars. Arch. Oral. Biol. 57, 790–795 (2012).
Lidral, A. C. & Reising, B. C. The role of MSX1 in human tooth agenesis. J. Dent. Res. 81, 274–278 (2002).
Sandhu, S. & Kaur, T. Radiographic evaluation of the status of third molars in the Asian-Indian students. J. Oral Maxillofac. Surg. 63, 640–645 (2005).
Celikoglu, M. & Kamak, H. Patterns of third-molar agenesis in an orthodontic patient population with different skeletal malocclusions. Angle Orthod. 82, 165–169 (2012).
Mok, Y. Y. & Ho, K. K. Congenitally absent third molars in 12 to 16 year old Singaporean Chinese patients: a retrospective radiographic study. Ann. Acad. Med. Singap. 25, 828–830 (1996).
Kazanci, F., Celikoglu, M., Miloglu, O. & Oktay, H. Third-molar agenesis among patients from the East Anatolian Region of Turkey. J. Contemp. Dent. Pract. 11, E033–E040 (2010).
Bailleul-Forestier, I., Molla, M., Verloes, A. & Berdal, A. The genetic basis of inherited anomalies of the teeth. Part 1: clinical and molecular aspects of non-syndromic dental disorders. Eur. J. Med. Genet. 51, 273–291 (2008).
Kajii, T. S., Sato, Y., Kajii, S., Sugawara, Y. & Iida, J. Agenesis of third molar germs depends on sagittal maxillary jaw dimensions in orthodontic patients in Japan. Angle Orthod. 74, 337–342 (2004).
Tan, S. P., van Wijk, A. J. & Prahl-Andersen, B. Severe hypodontia: identifying patterns of human tooth agenesis. Eur. J. Orthod. 33, 150–154 (2011).
Mitsiadis, T. A. & Luder, H. U. Genetic basis for tooth malformations: from mice to men and back again. Clin. Genet. 80, 319–329 (2011).
Ferguson, C. A., Tucker, A. S. & Sharpe, P. T. Temporospatial cell interactions regulating mandibular and maxillary arch patterning. Development 127, 403–412 (2000).
Wang, J., Jian, F., Chen, J., Wang, H., Lin, Y., Yang, Z. et al. Sequence analysis of PAX9, MSX1 and AXIN2 genes in a Chinese oligodontia family. Arch. Oral Biol. 56, 1027–1034 (2011).
Liang, J., Song, G., Li, Q. & Bian, Z. Novel missense mutations in PAX9 causing oligodontia. Arch. Oral Biol. 57, 784–789 (2012).
Hanihara, K. Racial characteristics in the dentition. J. Dent. Res. 46, 923–926 (1967).
Kanazawa, E., Sekikawa, M. & Ozaki, T. A quantitative investigation of irregular cuspules in human maxillary permanent molars. Am. J. Phys. Anthropol. 83, 173–180 (1990).
Acknowledgements
We are grateful to the individuals who participated in this study. This work was partly supported by a KAKENHI Grant-in-Aid for challenging Exploratory Research 25670881 (to TY), a MEXT-Supported Program for the Strategic Research Foundation at Private Universities, 2012–2017, and a Grant in Aid for Scientific Research on Innovative Areas “Genome Science” from the Ministry of Education, Culture, Sports, Science and Technology of Japan (No. 221S0002).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Haga, S., Nakaoka, H., Yamaguchi, T. et al. A genome-wide association study of third molar agenesis in Japanese and Korean populations. J Hum Genet 58, 799–803 (2013). https://doi.org/10.1038/jhg.2013.106
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2013.106