Abstract
Prader–Willi syndrome (PWS) is a genetic disorder caused by the absence of expression of the paternal copy of maternally imprinted genes in chromosome region 15q11–13. The genetic subtypes of PWS are classified into deletion (∼70%), maternal uniparental disomy (mUPD; 25–30%), imprinting center defects (3–5%) and rare unbalanced translocations. Recently, Matsubara et al. reported a significantly higher maternal age in a trisomy rescue (TR) or gamete complementation (GC) by nondisjunction at maternal meiosis 1 (M1) group than in a deletion group. In the present study, we try to confirm their findings in an ethnically different population. A total of 97 Korean PWS patients were classified into deletional type (n=66), TR/GC (M1) (n=15), TR/GC by nondisjunction at maternal meiosis 2 (n=2), monosomy rescue or postfertilization mitotic nondisjunction (n=4) and epimutation (n=2). Maternal ages at birth showed a significant difference between the deletion group (median age of 29, interquartile range (IQR)=(27,31)) and the TR/GC (M1) group (median age of 35, IQR=(31,38)) (P<0.0001). The relative birth frequency of the TR/GC (M1) group has substantially increased since 2006 when compared with the period before 2005. These findings support the hypothesis that the advanced maternal age at childbirth is a predisposing factor for the development of mUPD because of increased M1 errors.
Similar content being viewed by others
Introduction
Prader–Willi syndrome (PWS) was first described in 1956 by endocrinologists Prader, Labhart and Willi. This disease is characterized by neonatal hypotonia and feeding problems, childhood onset hyperphagia and obesity, short stature, hypogonadism, intellectual disabilities and behavioral problems.1, 2, 3 PWS is a genetic disorder that occurs in 1/10 000–1/25 000 live births and is caused by the absence of expression of the paternal copy of maternally imprinted genes in chromosome region 15q11–13.4, 5 The frequencies of different subtypes in PWS are usually given in the literature as 70% paternal interstitial deletion of the 15q11–13 region, 25–30% maternal uniparental disomy (mUPD) of chromosome 15, 3–5% imprinting center (IC) defects (microdeletion of PWS-IC, primary epimutation) and unbalanced translocations involving the PWS region.6, 7
The genetic findings in PWS can be explained by a concept known as genetic imprinting, where certain genes or groups of neighboring genes are expressed differently depending on the sex of the parent from which they were inherited.8 In general, mUPD is defined as the presence of both homologues of one chromosome from only mother, and is diagnosed using DNA polymorphism analysis of the proband and parental DNA. Four possible mechanisms of mUPD were proposed by Cassidy et al., including: (1) the formation of a trisomic zygote from a disomic egg with two maternal chromosome 15s plus a normal sperm, followed by subsequent loss of the paternal chromosome 15 (trisomy rescue, TR); (2) a disomic egg plus a nullisomic sperm without a chromosome 15, leading to a normal chromosome count (gamete complementation, GC); (3) a monosomic egg plus a nullisomic sperm producing a monosomic zygote followed by duplication of the maternal chromosome 15 (monosomy rescue, MR); and (4) postfertilization nondisjunction producing mosaicism for trisomic and monosomic cell lines, with subsequent duplication in the monosomic line (postfertilization mitotic nondusjunction, Mit).9 Two mechanisms can give rise to TR/GC: (1) nondisjunction of the homologous chromosome 15s during female meiosis 1 (M1); and (2) nondisjunction of the two sister (identical) chromatids during meiosis 2 (M2). The possibility of M1 error, in particular, has been correlated with increased maternal age because of a long-term meiotic arrest at prophase 1.10 Recently, Matsubara et al. reported a significantly higher maternal age in a TR/GC (M1) group than in a deletion group and a greater relative frequency of the TR/GC (M1) group in patients born in Japan since the year 2003 than in those born before 2002.11
In the present study, we aimed to confirm the hypothesis that the occurrence of mUPD caused by meiosis I error is positively correlated with maternal age at birth. In total, 97 Korean PWS patients whose parental age was available were classified into genetic subtype in detail. In addition, we examined the effects of maternal age on the distribution of genetic subtypes in PWS by comparing the maternal age of PWS patients at birth according to their genetic subtypes.
Materials and methods
Patients with PWS
This study protocol was approved by the Institute Review Board Committees at the Samsung Medical Center (IRB No. SMC 2010-11-005-002). Informed consent was confirmed by the IRB. The study included 97 Korean PWS patients (median birth year of 2001, interquartile range (IQR)=(1998, 2007), male:female=58:39 patients) who revealed hypermethylated PWS-IC, which was confirmed by methylation analysis. All patients met the consensus clinical criteria for PWS. Data on the parental age at childbirth had been collected from all patients by interviews with their parents when the patients visited the outpatient clinic, as scheduled from January 2011 to October 2011. When further molecular tests were necessary to classify the genetic subtype, those tests were performed after obtaining the written consent from the parents.
Flow of the molecular studies
Diagnosis of all PWS patients was confirmed by methylation-specific PCR, as previously described.12 We then performed fluorescence in situ hybridization analysis with commercial probes (LSI SNRPN SpectrumOrange/CEP15 D15Z1 SpectrumGreen/PML SpectrumOrange, Vysis, Downers Grove, IL, USA) in order to detect the interstitial deletion of the 15q11–13 region.13 In addition, microsatellite analysis was undertaken using a Devyser UPD-15 diagnostic kit (Devyser AB, Hagersten, Sweden) as well as additional pericentromeric markers, including D15S541, D15S542, D15S1035 and D15S11, in order to confirm the mUPD when blood samples of parents were available (Table 1). The Devyser UPD-15 kit includes nine tetra-repeat short tandem repeat markers that are specific for chromosome 15, and four of these short tandem repeat markers are located within the 15q11–13 deletion region. Finally, we performed multiplex ligation-dependent probe amplification (MLPA) analysis and chromosomal microarray in the patients with neither 15q11–13 deletion nor mUPD, in order to identify the IC deletion. For MLPA analysis, we utilized a commercially available MLPA probe mix (ME028) (MRC-Holland, Amsterdam, The Netherlands), as previously described.14 For chromosomal microarray, we used an Affymetrix Cytogenetics Whole-Genome 2.7M Array (Affymetrix Inc., Santa Clara, CA, USA), as previously described.15
Classification of genetic subtypes in the patients with PWS
We classified the patients with PWS into seven subgroups by the underlying genetic mechanisms. The mUPD patients were divided into three subgroups, based primarily on the results of microsatellite analysis, with the assumption of no recombination between the centromere and the four pericentrometric loci within the 15q11–13 region, as described previously.16 In the present study, if a patient showed heterodisomy for at least one of the four adjacent pericentromeric microsatellite loci (D15S541, D15S542, D15S1035 and D15S11), the patient was classified as having TR/GC type mUPD caused by M1 non-disjunction (TR/GC (M1) group). Second, a patient who showed isodisomy for the pericentromeric microsatellite loci and heterodisomy for at least one middle to distal microsatellite loci was classified as having TR/GC type mUPD through M2 non-disjunction (TR/GC (M2) group). Lastly, when a patient revealed isodisomy for all the microsatellite loci, the patient was classified as having MR/Mit type mUPD (MR/Mit group). If patients who had biparental pattern in the results of microsatellite analysis revealed no microdeletions by MLPA/chromosomal microarray, those patients were classified as having primary epimutation.
Statistical analysis
Comparisons were made of parental ages between two genetic subtypes (Deletion vs TR/GC (M1)) and between two different time periods (up until the year 2005 and since the year 2006), and the relative frequency of each group between the two time periods. Tests of statistical significance included the Mann–Whitney test for the median age, Spearman’s correlation analysis for the correlation between parental ages, the χ2-test for the relative frequency between the two genetic subtypes (Deletion vs TR/GC (M1)), and the Mann–Whitney test for the relative frequency between the two different time periods (before the year 2005 and since the year 2006). P<0.05 was considered significant.
Results
Classification of genetic subtypes in the patients with PWS
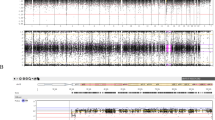
The information of parental age at birth was collected for all 97 patients, who consisted of 66 patients with deletions and 31 patients without deletions. Microsatellite genotyping was then carried out in 23 of the 31 patients without deletions, which classified 15 patients as a TR/GC (M1) group, 2 patients as a TR/GC (M2) group and 4 patients as a MR/Mit group. The remaining eight patients, who could not undergo further studies, were classified as non-deletion types. No findings were indicative of segmental isodisomy or mosaicism. MLPA and chromosomal microarray were performed in the two non-mUPD patients, but identified no microdeletion affecting the PWS-IC. These two patients were ultimately classified as a primary epimutation group. All of these results are presented in Figure 1.
Trend of maternal age
Korean women have shown a recent trend of getting married at a later age and they subsequently have their children later in life. The Korean Statistical Information Service reported that the mean maternal age at the 1st child’s birth was 27.55 years in 1993, whereas it was 31.26 years in 2010. In addition, the birthrate for women 25–29 years of maternal age was highest among all age groups, and therefore was higher than that of women 30–34 years of maternal age, up until 2005. However, since 2006, the birthrate of two age groups has been reversed, and birthrate of women 30–34 years of maternal age is now the highest. Therefore, we hypothesized that the incidence of the TR/GC (M1) form of PWS would have increased in Korea since 2006.
Analysis of parental ages between the deletion and the TR/GC (M1) group
We compared parental ages only between the Deletion and the TR/GC (M1) group, because other groups were too small to compare statistically. Comparison of maternal ages revealed a significant difference between the Deletion (median age of 29, IQR=(27,31)) and the TR/GC (M1) groups (median age of 35, IQR=(31,38)) (Mann–Whitney test, P<0.0001). Paternal ages showed a similar tendency, with a significant difference between the Deletion (median age of 31.5, IQR=(29,34)) and the TR/GC (M1) groups (median age of 36 years, IQR=(32,40)) (Mann–Whitney test, P=0.003) (Figure 1a). A significant correlation was observed between maternal and paternal ages in the Deletion group (r=0.742; P<0.0001, Spearman’s correlation analysis), and a moderate correlation was found between maternal and paternal ages in the TR/GC (M1) group (r=0.689; P=0.005, Spearman’s correlation analysis) (Figure 2b).
Parental ages at childbirth according to PWS genetic subtypes. (a) Parental ages in each genetic subgroup. (b) Correlation between maternal and paternal ages at childbirth. Significant correlation is observed in the Deletion group, and moderate correlation in the TR/GC (M1) group. (c) Relative frequency of TR/GC (M1) was markedly increased in the 32 patients born since 2006 (N=17, 10, 2 and 3 for deletion, TR/GC (M1), MR/Mit and non-deletions groups, respectively), compared with the 65 patients born before 2005 (N=49, 5, 2, 2, 2 and 5 for Deletion, TR/GC (M1), TR/GC (M2), MR/Mit, epimutation and non-deletions groups, respectively). A full color version of this figure is available at the Journal of Human Genetics journal online
The relative frequency of the deletion and the TR/GC (M1) group according to the time period
The relative frequency of each group markedly differed when comparing the 65 patients born up until 2005 and the 32 patients born since 2006 (Figure 2c). A TR/GC (M1) was indicated in 5 of the 65 patients born before 2005, and 5 non-deletion type patients were born following 2005. Thus, the TR/GC (M1) group accounted for at least 5 and up to 10 of the 65 patients born until the year 2005, and at least 10 and up to 13 of the 32 patients born since the year 2006. This difference in relative frequency of TR/GC (M1) was assessed as statistically significant, as the P-values were <0.001 for 5/65 versus 10/32, and 0.008 for 10/65 versus 13/32 (χ2-test). For all patients, both maternal ages and paternal ages at birth significantly differed between the two time periods (Mann–Whitney test, maternal age, P=0.003; paternal age, P=0.002).
Discussion
In total, 97 patients with PWS were classified into several detailed genetic subgroups by stepwise genetic analysis: deletions in 66 patients (68%), mUPD in 21 patients (21.6%), non-deletion in 8 patients (8.2%) and primary epimutation in 2 patients (2.1%). Microdeletion of PWS-IC, which can be inherited in the next generation, was not found. The actual frequency of mUPD could be as high as 28 patients (29.9%) because most of the 8 patients with non-deletions might be classified as mUPD, considering the rare prevalence of the other remaining causes. No coexistence was found among siblings who had PWS in our study. The proportions of deletions, mUPD, and other rare causes found in our study were similar to those reported in previous studies.3, 11 The subgroups of mUPD were classified into TR/GC (M1), TR/GC (M2) and MR/Mit groups by microsatellite analysis, and the TR/GC (M1) type mUPD was the most common (15 out of 21 patients).
At present, little is known about the factors that influence the distribution of genetic subtypes in PWS. Deletions, unbalanced translocations and imprinting defects appear to be random genetic accidents—one report has suggested that paternal contact with hydrocarbons could be important, but this has never been confirmed.17 An association of mUPD with increased maternal age has also been suggested.3, 18 The births of babies with trisomies, as in Down’s syndrome, are well known to occur more frequently in older mothers than in their younger counterparts.19, 20
In the present study, the maternal ages at birth were significantly higher in the TR/GC (M1) group than in the Deletion group, which was free of any maternal age effect. These results are consistent with recent Japanese study reported by Matsubara et al.11 The lack of any significant difference in the maternal ages between the maternal age-dependent TR/GC (M1) group and the other maternal age-independent subgroups could be due primarily to the small number of patients in the other subgroups. The relative frequency of the TR/GC (M1) group has significantly increased since 2006, when delayed childbearing age became obvious in the Korean population. The advanced maternal ages at birth since the year 2006 were primarily associated with the high frequency of the TR/GC (M1) group.
The traditional estimate of the proportion of mUPD babies is 25–30% and the prevalence of PWS patients is 1:10 000–1:25 000 live births. This gives a risk of 1:33 000–1:100 000 for giving birth to a mUPD child. One previous study showed that the risk rose exponentially with mother’s age, as in Down’s syndrome, reaching 1:3400 for women older than 40 years.21 The trend toward more PWS mUPD births in Korea supposedly reflects the country’s advanced economy, the trend toward delaying marriage, and a trend toward having families later in life. If we assume that the numbers of PWS cases due to deletions are not decreasing, we would expect a small increase in the birth incidence of PWS, given the increase in the proportion of those with mUPD because of increasing proportions of older mothers.
We did not include any patients without fluorescence in situ hybridization results. Most patients included in our study have been followed up actively at our clinic. Other possible explanations of recent increases in mUPD should be considered in addition to maternal age; for example, undiagnosed mUPD cases in older patients, sampling error, the possibility of some environmental variable causing genetic errors and the use of reproductive technologies. We investigated the frequency of artificial fertilization in each genotype, but only one patient with a deletion and one patient with TR/GC (M1) were born by in vitro fertilization in our study. Therefore, any influence of artificial fertilization on the genotype could not be estimated because of too small a number. Further study will be needed to determine whether assisted reproductive technologies might be a potential factor involved in the molecular cause of genetic abnormalities.
In summary, we confirmed an influence of the trend toward later marital age on the genetic subtype distribution of the PWS population in Korea. The information reported here may be helpful in the classification of the (epi)genetic type in PWS and for genetic counseling of parents. Further study of the correlation between (epi)genetic type and clinical phenotype will be needed in order to offer more information about clinical courses and prognosis.
References
Cassidy, S. B. Prader-Willi syndrome. J. Med. Genet. 34, 917–923 (1997).
Holm, V. A., Cassidy, S. B., Butler, M. G., Hanchett, J. M., Greenswag, L. R., Whitman, B. Y. et al. Prader-Willi syndrome: consensus diagnostic criteria. Pediatrics 91, 398–402 (1993).
Cassidy, S. B. & Driscoll, D. J. Prader-Willi syndrome. Eur. J. Hum. Genet. 17, 3–13 (2009).
Donaldson, M. D., Chu, C. E., Cooke, A., Wilson, A., Greene, S. A. & Stephenson, J. B. The Prader-Willi syndrome. Arch. Dis. Childhood 70, 58–63 (1994).
Kaplan, J., Fredrickson, P. A. & Richardson, J. W. Sleep and breathing in patients with the Prader-Willi syndrome. Mayo Clinic Proc. Mayo Clinic 66, 1124–1126 (1991).
Ledbetter, D. H., Riccardi, V. M., Airhart, S. D., Strobel, R. J., Keenan, B. S. & Crawford, J. D. Deletions of chromosome 15 as a cause of the Prader-Willi syndrome. N. Engl. J. Med. 304, 325–329 (1981).
Nicholls, R. D., Knoll, J. H., Butler, M. G., Karam, S. & Lalande, M. Genetic imprinting suggested by maternal heterodisomy in nondeletion Prader-Willi syndrome. Nature 342, 281–285 (1989).
Driscoll, D. J., Waters, M. F., Williams, C. A., Zori, R. T., Glenn, C. C., Avidano, K. M. et al. A DNA methylation imprint, determined by the sex of the parent, distinguishes the Angelman and Prader-Willi syndromes. Genomics 13, 917–924 (1992).
Cassidy, S. B., Lai, L. W., Erickson, R. P., Magnuson, L., Thomas, E., Gendron, R. et al. Trisomy 15 with loss of the paternal 15 as a cause of Prader-Willi syndrome due to maternal disomy. Am. J. Hum. Genet. 51, 701–708 (1992).
Jones, K. T. Meiosis in oocytes: predisposition to aneuploidy and its increased incidence with age. Hum. Reprod. Update 14, 143–158 (2008).
Matsubara, K., Murakami, N., Nagai, T. & Ogata, T. Maternal age effect on the development of Prader-Willi syndrome resulting from upd(15)mat through meiosis 1 errors. J. Hum. Genet. 56, 566–571 (2011).
Kubota, T., Das, S., Christian, S. L., Baylin, S. B., Herman, J. G. & Ledbetter, D. H. Methylation-specific PCR simplifies imprinting analysis. Nat. Genet. 16, 16–17 (1997).
Kim, H. J., Cho, H. J., Jin, D. K., Kwon, E. K., Ki, C. S., Kim, J. W. et al. Genetic basis of Prader-Willi syndrome in Korea: less uniparental disomy than has been recognized? Clin. Genet. 66, 368–372 (2004).
Procter, M., Chou, L. S., Tang, W., Jama, M. & Mao, R. Molecular diagnosis of Prader-Willi and Angelman syndromes by methylation-specific melting analysis and methylation-specific multiplex ligation-dependent probe amplification. Clin. Chem. 52, 1276–1283 (2006).
Harvard, C., Strong, E., Mercier, E., Colnaghi, R., Alcantara, D., Chow, E. et al. Understanding the impact of 1q21.1 copy number variant. Orphanet. J. Rare Dis. 6, 54 (2011).
Robinson, W. P., Kuchinka, B. D., Bernasconi, F., Petersen, M. B., Schulze, A., Brondum-Nielsen, K. et al. Maternal meiosis I non-disjunction of chromosome 15: dependence of the maternal age effect on level of recombination. Hum. Mol. Genet. 7, 1011–1019 (1998).
Akefeldt, A., Anvret, M., Grandell, U., Nordlinder, R. & Gillberg, C. Parental exposure to hydrocarbons in Prader-Willi syndrome. Dev. Med. Child Neurol. 37, 1101–1109 (1995).
Cassidy, S. B., Forsythe, M., Heeger, S., Nicholls, R. D., Schork, N., Benn, P. et al. Comparison of phenotype between patients with Prader-Willi syndrome due to deletion 15q and uniparental disomy 15. Am. J. Med. Genet. 68, 433–440 (1997).
Warburton, D. Biological aging and the etiology of aneuploidy. Cytogenet. Genome Res. 111, 266–272 (2005).
Sherman, S. L., Freeman, S. B., Allen, E. G. & Lamb, N. E. Risk factors for nondisjunction of trisomy 21. Cytogenet. Genome Res. 111, 273–280 (2005).
Robinson, W. P., Langlois, S., Schuffenhauer, S., Horsthemke, B., Michaelis, R. C., Christian, S. et al. Cytogenetic and age-dependent risk factors associated with uniparental disomy 15. Prenat. Diagn. 16, 837–844 (1996).
Acknowledgements
The study was supported by the Korea Healthcare Technology R&D Project funded by the Ministry of Health, Welfare and Family Affairs, Republic of Korea (grant number A080588), the Samsung Biomedical Research Institute (grant number C-A9-240-3) and the In-Sung Foundation for Medical Research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yoon Cho, S., Ki, CS., Bae Sohn, Y. et al. The proportion of uniparental disomy is increased in Prader–Willi syndrome due to an advanced maternal childbearing age in Korea. J Hum Genet 58, 150–154 (2013). https://doi.org/10.1038/jhg.2012.148
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2012.148
Keywords
This article is cited by
-
A female with typical fragile-X phenotype caused by maternal isodisomy of the entire X chromosome
Journal of Human Genetics (2020)
-
Exploring autism symptoms in an Australian cohort of patients with Prader-Willi and Angelman syndromes
Journal of Neurodevelopmental Disorders (2018)
-
Prenatal molecular testing for Beckwith–Wiedemann and Silver–Russell syndromes: a challenge for molecular analysis and genetic counseling
European Journal of Human Genetics (2016)
-
Imprinting methylation errors in ART
Reproductive Medicine and Biology (2014)
-
Advanced maternal age at childbirth and the development of uniparental disomy. A commentary on the proportion of uniparental disomy is increased in Prader–Willi syndrome due to an advanced maternal childbearing age in Korea
Journal of Human Genetics (2013)