Abstract
In the course of our screening for inhibitors of lipopolysaccharide (LPS) binding to cellular receptor CD14, potent inhibitory activity was detected in the cultured broth of Pedobacter sp. SANK 72003. Three active compounds, pedopeptin A, B and C, were isolated from the broth and their structures were elucidated by physicochemical and spectral data to be new cyclic depsipeptides.
Similar content being viewed by others
Introduction
Sepsis is a severe symptom caused by overwhelming infections from various types of bacteria, and septic shock may be a life-threatening event in clinical situations. Gram-negative bacteria have lipopolysaccharide (LPS) in their cell wall. LPS is well known as a bacterial endotoxin and is a key component of septic shock. A reaction is started by LPS binding to its receptors, and the succeeding overexpression of inflammatory cytokines triggers the symptoms. Therefore, a compound that inhibits the LPS binding to cellular receptor CD14,1 may be useful as a therapeutic agent for sepsis and septic shock.
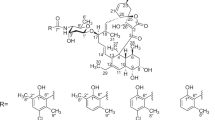
In the course of our screening for inhibitors of LPS binding to CD14, potent activity was detected in the cultured broth of Pedobacter sp. SANK 72003, and three active compounds, pedopeptin A (1), B (2) and C (3) were isolated (Figure 1).
In this paper, we report the taxonomy of the producing organism, fermentation, isolation, physicochemical properties and structural elucidations of pedopeptins. Details of the biological activities of pedopeptins are reported independently.2
Results
Taxonomy of producing organism
The strain SANK 72003 was a Gram-negative, rod-shaped and had no motility. No spore was formed. Colonies of SANK 72003 on nutrient agar were smooth, creamy white in color, circular and entire-edged. This strain was positive in catalase, oxidase and DNase production tests, and was fermentative in an O-F test. The growth temperature range was 10–31 °C. A BLAST search of 16S rDNA sequence showed the highest similarities (99%) with Pedobacter himalayensis HHS22T (AJ583425). Phylogenetic analysis of 16S rDNA sequences based on the neighbor-joining method suggested that SANK 72003 was clustered in the genus Pedobacter. According to Bergey’s Manual of Systematic Bacteriology, the morphological and physiological characteristics of SANK 72003 also resembled those of the genus Pedobacter. Thus, this strain was identified as Pedobacter sp. SANK 72003 (Figure 2). The strain SANK 72003 was deposited in the International Patent Organism Depositary, National Institute of Advanced Industrial Science and Technology, Ibaraki Prefecture, Japan with the accession number FERM BP-08549.
Phylogenetic analysis of 16S rDNA sequences based on the neighbor-joining method indicating the taxonomic position of the strain SANK 72003. Bootstrap values (%) are indicated at the branches from 1000 replications. The scale bar below the phylogenetic tree indicates five nucleotide substitutions per 100 nucleotides.
Fermentation
One-loopful of cells of Pedobacter sp. SANK 72003 was transferred from an agar slant into a 2-l Erlenmeyer flask containing 300 ml of a sterilized primary seed medium composed of nutrient broth (Eiken Kagaku) 1.8%. The flask was incubated at 28 °C for 47 h on a rotary shaker at 210 r.p.m. The resulting primary seed culture (300 ml) was transferred into a 30-l jar fermentor containing 15 l of a production medium, which was the same as the primary seed medium. The fermentation was carried out at 28 °C for 70 h with agitation (100–220 r.p.m.) and aeration (7.5 l min−1).
Isolation
The active compounds are detected by the LPS-CD14 binding assay2 or HPLC.
The cultured broth (60 l, four fermentations) was centrifuged. The pH of the supernatant was adjusted to 8.0 with 6N HCl and was subjected to a Diaion HP-20 column (7.2 l). After washing the column with water (21 l), 50% aq. acetone (21 l) and 60% aq. acetone–0.1% formic acid (5.4 l), the active compounds were eluted with 60% aq. acetone–0.1% formic acid (8.0 l). The eluate was concentrated, and lyophilized to give a powder (6.57 g).
The powder was dissolved in MeCN–0.3% triethylamine-phosphate (TEAP) buffer at pH 3.0 in the ratio of 1:9 (800 ml) and was subjected to an ODS open column (Cosmosil 140C18-OPN, 2.4 l). The chromatography was performed by stepwise elution using MeCN–0.3% TEAP buffer at pH 3.0 in the ratio of 1:3 (5 l), 3:7 (8 l), 7:13 (5.5 l) and 2:3 (3 l). The ratio 7:13 and 2:3 eluates were fractionated (300 ml) and then analyzed for the compounds in each fraction by HPLC using an ODS column (SYMMETRY C18, 4.6 i.d. × 150 mm, Waters (Milford, MA, USA)).
Isolation of Pedopeptin A (1)
The part of the eluate at a ratio of 7:13 containing 1, was concentrated to remove MeCN, and the pH was adjusted to 6.0 with 10% aq. NaOH. It was then subjected to an HP-20 column (300 ml) for desalting. After washing the column with water (1.5 l), 1 was eluted with 60% aq. acetone–0.1% formic acid (900 ml). The fraction containing 1 was concentrated and lyophilized to give pure 1 as a white powder (1.3 g).
Isolation of Pedopeptin B (2)
The part of the eluate at a ratio of 2:3 containing 2, was concentrated to remove MeCN, and the pH was adjusted to 6.0 with 10% aq. NaOH. It was then desalted in the same manner as described above and lyophilized to give pure 2 as a white powder (98 mg).
Isolation of Pedopeptin C (3)
The part of the eluate at a ratio of 7:13 containing 3, was concentrated to remove MeCN, and the pH was adjusted to 6.0 with 10% aq. NaOH. It was then desalted, and lyophilized to give a partially purified substance (1.15 g). The substance was re-dissolved in 50 ml of MeCN–0.3% TEAP buffer at pH 3.0 (1:3) and was subjected to a Cosmosil column (100 ml). The chromatography was performed by stepwise elution using MeCN–0.3% TEAP buffer at pH 3.0 in the ratio 1:3 (320 ml) and 3:7 (600 ml). The part of the ratio 3:7 containing 3 was concentrated to remove MeCN and the pH was adjusted to 6.0 with 6 N NaOH. It was then desalted as described above and lyophilized to give 3 as a white powder (132 mg).
Physicochemical properties
The physicochemical properties of pedopeptins are summarized in Table 1.
Structural elucidation
Structural elucidation of pedopeptin A (1)
The structural elucidation was mainly focused on 1. The 1H and 13C NMR spectral data of 1 as summarized in Table 2 showed the typical features of a peptide antibiotic. In the 1H NMR spectrum, signals from α-methine of the amino-acid residues (δ 4.1–5.8) and the phenyl group (δ 7.1–7.4) were observed. Although three carbon signals were overlapped in the 13C NMR spectrum, all 53 carbons were classified into 9 × methyl, 12 × methylene, 18 × methine and 14 quaternary carbons by their DEPT and HSQC spectra.
Amino-acid analysis (AAA) of the acid hydrolysate detected one mole of aspartic acid, two moles of leucine, one mole of phenylalanine and unidentified amino acids. GC-MS analysis of the trimethylsilyl derivatives of the acid hydrolysate not only supported the AAA result, but also proved that two of the unidentified amino acids were diaminopropionic acid (Dap) and diaminobutyric acid (Dab).
The IR spectral data indicated the presence of ester groups (1732 cm−1). In the ESI-MS analysis, mild alkaline hydrolysate of 1 (4, Figure 3) showed a protonated ion (m/z 1133.6), which had 18 mass units larger than that of 1. These data indicated that 4 was a linear peptide from 1 and that 1 was a cyclic depsipeptide.
As the signals in the 2D NMR spectra of 1 were crowded, partial structures were elucidated by analyses of the partially digested products of 1; that is, 4 was digested with chymotrypsin to provide two products (5 and 6, Figure 3).
GC-MS analysis of the acid hydrolysate of 5 showed the existence of Phe and Dap. By several NMR techniques, including DQFCOSY, HSQC and HMBC, 5 was elucidated to be a peptide consisting of Phe, Dap and a 3-hydroxy-7-methyl octanoic acid moiety.
Structure determination of 6 was carried out in the same way as 5. The GC-MS analysis of the acid hydrolysate of 6 showed the existence of Asp, Leu, Dap, Dab and unidentified amino acids. The partial structure of 6, except for the bond between Dab and 2-amino-2-butenoic acid (dehydrobutyrine, Dhb), was elucidated by several NMR techniques that were the same as 5. In addition, the unidentified amino acids were proved to be β-hydroxyvaline and Dhb by the NMR analysis. The bond between Dab and Dhb was elucidated by the product ions (m/z 561 and 644) in the collision-induced dissociation spectra of 6, as shown in Figure 4.
Chymotrypsin digestion is known to occur at the carboxy terminus of Phe. The C-terminal amino acid of 5 assigned to Phe was considered to connect with Dab, which is the amino terminus of 6, because a product ion (m/z 644) was observed in the collision-induced dissociation spectra of 4 (Figure 4). Observed C-H long-range couplings between H-2 of Dab residue (δ 4.56) and C-1 of Phe residue (δ 174.1) in the band-selective HMBC spectrum3 of 1 also supported this result. Thus, the structure of 4 was revealed as shown in Figure 3.
In the band-selective HMBC spectrum of 1, C-H long-range coupling between H-3 of a fatty acid moiety (δ 5.19) and a carbonyl carbon of Asp residue (δ 173.2) was also observed. However, it was still uncertain which carboxyl group in the Asp residue was condensed with the hydroxyl group of the fatty acid. Because an esteric carbonyl is more easily reduced than a carboxylic one by lithium borohydride, selective reduction was applied for the determination. As the reduction was not carried out by using 1 itself, an acetylated derivative of 1 (7) was used as a substrate. By this reaction, C-1 of the Asp residue was reduced to a methylene. This result clearly indicated that the carbonyl at the C-1 position of the Asp residue took part of the ester bond, and the planar structure of 1 was elucidated as shown in Figure 1.
The geometry of Dhb in 1 was determined by the chymotrypsin-digested product of 7 (9) because the NMR spectra of 1 and 6 in dimethyl sulfoxide (DMSO)-d6 did not give a good resolution and that of 7 was crowded. In the NOE difference spectrum, observed NOE between NH-2 of Leu-1 (δ 8.47) and H-4 of Dhb (δ 1.77) indicated that the stereochemistry of Dhb was E.
The stereochemistry of each amino-acid residue of 1 was determined by the advanced Marfey's method.4 Acid hydrolysates of 1 were coupled with L- or D-FDLA (1-fluoro-2,4-dinitrophenyl-5-leucine amide), and analyzed by LC-MS. By comparing each authentic amino-acid standard, the chiralities of Asp, Phe, Dab and β-hydroxyvaline were determined to be L configurations. As well, the two leucines and two Daps were mixtures of D and L isomers. To determine the configuration of the two Daps, 5 and 6 were hydrolyzed, coupled with FDLA and analyzed independently. As a result, each FDLA derivative exhibited two peaks assigned to be D and L derivatives.
Structural elucidation of other homologs
Structural elucidation of the homologs, pedopeptin B (2) and C (3) were carried out in the same way as pedopeptin A (1). The 1H and 13C NMR spectral data of 2 and 3 are summarized in Tables 3 and 4, respectively.
The molecular formula of 2 had one less oxygen atom than that of 1. AAA of the acid hydrolysate of 2 gave the same amino acids as 1 except for β-hydroxyvaline, and one mole of Val was observed instead. By mass spectral and NMR spectral data, the structure of 2 had a Val instead of β-hydroxyvaline of 1.
The molecular formula of 3 suggested that 3 is a demethyl analog of 1. AAA of the acid hydrolysate of 3 showed the existence of the same amino acids as 1. By mass spectral and NMR spectral data, 3 had a normal β-hydroxyfatty acid elucidated as 3-hydroxy-n-octanoic acid.
Discussion
In the course of our screening for inhibitors of LPS binding to CD14, pedopeptin A (1), B (2) and C (3) were isolated from the cultured broth of Pedobacter sp. SANK 72003. The structures of the compounds were elucidated by physicochemical and spectral data analysis, and they were determined to be new cyclic depsipeptides.
The genus, Pedobacter was dissociated from Sphingobacterium in 1998.5 Although Pedobacter is a relatively newly established genus, it has often been reported to have an interesting enzyme production profile.6, 7, 8 Pedopeptins are the first bioactive low MW compounds reported from Pedobacter.
NMR analysis of the pedopeptins was performed in DCl/D2O (1 and 3) or methanol-d4 (2), because other solvents did not give good solubility and resolution. For example, when 1 was dissolved in DMSO, it did not give good resolution. In the geometry determination study of Dhb, several modifications were needed for the observation of NMR signals.
According to the stereochemistry determination of Dhb in the case of Sch 20562,9 the olefin signal of the (Z)-Dhb appears at lower field (δ 6.48) than that of the (E)-Dhb (δ 5.90) in DMSO-d6. The same phenomena had also been reported in other Dhb-containing compounds.10, 11 These reports also supported our determination that the geometry of Dhb in pedopeptins is E (δ 5.63–5.65).
Regarding the stereochemistry of the two leucines and two Daps, both D and L isomers were observed. In the case of leucines, it was difficult to determine which one had L-configuration, as they were included in the same digested-peptide fragment. In the case of Daps, it is known to racemize under the standard acidic hydrolysis conditions.12, 13, 14 Our data also suggested that both of Dap-1 and Dap-2 existed as racemates. The other supporting data is necessary for the final determination.
Polymyxins,15 which are known as antibiotics and LPS binders, prevent many toxic effects due to LPS.16, 17 They also showed potent activities in our assay system. Polymyxins are not cyclic depsipeptides, but are cyclic peptides including some basic amino acids and a long acyl chain. Although the acyl moiety of polymyxins was not indispensable for LPS binding, the length of the chain affected the strength of the binding affinity.18 The Dab residues are also important for the binding affinity.19 Although the structures of pedopeptins are different from those of polymyxins, they have three primary amines and a fatty acyl chain in their structures. Pedopeptins are also considered to be LPS binders. Pedopeptins and polymyxins may also have the same mechanism of the inhibition originating from their basicity and a fatty acyl moiety in the molecule.
Experimental section
General experimental procedures
IR spectra were obtained with a JASCO FT/IR-610 spectrometer. UV spectra were recorded on a Shimadzu UV-265FW spectrometer. NMR spectra were recorded on Bruker Biospin AVC500 spectrometers equipped with C-H dual and TXI cryogenic probes, respectively. High-resolution mass spectra were recorded on a Micromass liquid chromatography time spectrometer. GC-MS spectra were recorded on a HP6890 Series GC System equipped with a column (DB-5, 0.25 mm i.d. × 15 m, Agilent Technologies (Santa Clara, CA, USA)) and a 5973 Mass Selective Detector.
Producing organism
The bacterial strain SANK 72003 was isolated from the rhizosphere soil sample collected at Sanada-town, Nagano Prefecture, Japan. The strain was routinely grown on nutrient agar at 28 °C for 2 days. The physiological characteristics were determined using standard protocols.20 16S rDNA was amplified by PCR and the amplified fragment was sequenced directly. Multiple alignments were performed by the program Clustal W.21 Phylogenetic analysis was performed by the neighbor-joining method.22
Amino-acid analysis
1 (100 μg) was completely hydrolyzed with 6 N HCl (400 μl) at 105 °C for 15 h. The hydrolysate was concentrated to dryness and dissolved in 0.02 N HCl, and applied to a HITACHI L-8500 Amino-Acid Analyzer.
Analysis of amino acid by GC-MS
1 (1.0 mg) was completely hydrolyzed in the same way as described above. To the solution of the hydrolysate in pyridine (100 μl) was added bis-(trimethylsilyl)trifluoroacetamide (Sigma-Aldrich, 100 μl). The mixture was diluted with pyridine and applied to a GC-MS. Each authentic amino acid was also prepared and analyzed in the same way.
Alkaline hydrolysate (4) of 1
1 (20 mg) was dissolved in 0.02N NaOH (20 ml) and stirred at room temperature for 22 h. The mixture was charged to an HP-20 column (5 ml) for desalting. After washing the column with water (15 ml), the absorbed lysate was eluted with 60% aq. acetone–0.1% formic acid (15 ml). The eluate was concentrated and lyophilized. Finally, the alkaline hydrolysate was purified by preparative HPLC using an ODS column (SYMMETRY C18, 19 i.d. × 100 mm, Waters). The chromatography was performed with 37% MeCN in aq. 5 mM HCOONH4-HCOOH (pH 3.0) at a flow rate of 6 ml min−1 to give 10.4 mg of 4.
Chymotrypsin digestion of 4
Digestion
4 (7.6 mg) was dissolved in the solvent, which composed of 0.04 M Tris–HCl buffer at pH 8.0 (5 ml). Chymotrypsin (1 mg) was dissolved in 0.001N HCl (1 ml). After pre-incubation at 37 °C for 15 min, the chymotrypsin solution (200 μl) was added to the substrate solution. The mixture was incubated at 37 °C for 2.5 h. The reaction was quenched by the addition of 50 mM aq. HCOONH4-HCOOH (pH 3.0).
Purification
The digestion described above gave two products (5 and 6). Each product was isolated using a Sep-pak C18 Plus cartridge (Waters). The reaction mixture was applied to the cartridge. After washing with water (5 ml), the absorbents were eluted stepwise using MeCN-5 mM aq. HCOONH4-HCOOH at pH 3.0 in the ratio of 1:9, 1:4, 3:7 and 1:1 (5 ml, each).
Compound 5
The eluate at a ratio of 3:7 containing 5 was concentrated and lyophilized to give a white powder of 5 (2.7 mg). ESI-MS (m/z): 408 (M+H)+. 1H NMR (500 MHz, MeOD, TMS (internal standard)): δ 0.89 (3H, d, 6.6 Hz), 0.89 (3H, d, 6.6 Hz), 1.20 (2H, m), 1.33 (1H, m), 1.39–1.49 (3H, overlapped), 1.55 (1H, m), 2.28 (1H, dd, 9.0, 14.3 Hz), 2.35 (1H, dd, 3.9, 14.3 Hz), 2.91 (1H, dd, 9.9, 14.3 Hz), 3.21 (2H, m), 3.30 (1H, overlapped), 3.95 (1H, m), 4.49 (1H, dd, 4.0, 9.7 Hz), 4.63 (1H, t, 6.1 Hz), 7.17 (1H, m), 7.22–7.29 (4H, overlapped). 13C NMR (125 MHz, MeOD, TMS (internal standard)): δ 23.0, 23.0, 24.5, 29.2, 38.7, 38.7, 40.1, 42.5, 44.5, 52.1, 58.0, 69.7, 127.5, 129.4, 129.4, 130.3, 130.3, 139.8, 170.1, 174.5, 178.2.
Compound 6
The eluate at ratios of 1:9 and 1:4 containing 6 were combined, concentrated and lyophilized to give a white powder of 6 (4.4 mg). ESI-MS (m/z): 744 (M+H)+. 1H NMR (500 MHz, D2O, HOD δ 4.80 (internal standard)): δ 0.86 (3H, d, 5.7 Hz), 0.92 (3H, d, 6.1 Hz), 0.96 (3H, d, 6.1 Hz), 0.94 (3H, d, 6.4 Hz), 1.26 (3H, s), 1.30 (3H, s), 1.51–1.62 (2H, overlapped), 1.63–1.81 (4H, overlapped), 1.83 (3H, d, 7.2 Hz), 2.32 (2H, m), 2.59 (1H, dd, 9.4, 15.6 Hz), 2.75 (1H, dd, 3.7, 15.6 Hz), 3.21 (2H, t, 8.1 Hz), 3.44 (1H, dd, 8.1, 13.2 Hz), 3.59 (1H, dd, 6.6, 13.2 Hz), 4.22 (1H, s), 4.22 (1H, overlapped), 4.32 (1H, dd, 2.9, 11.2 Hz), 4.45 (1H, dd, 3.7, 9.4 Hz), 4.59 (1H, dd, 4.6, 11.0 Hz), 4.72 (1H, t, 7.2 Hz), 5.94 (1H, q, 7.2 Hz). 13C NMR (125 MHz, D2O, 1,4-dioxane δ 67.2 (internal standard)): δ 13.3, 20.3, 21.0, 22.8, 23.1, 24.9, 25.0, 25.9, 26.6, 29.1, 35.9, 39.6, 39.6, 39.9, 40.5, 51.1, 51.7, 52.6, 53.6, 54.2, 62.4, 72.1, 124.3, 129.0, 167.5, 168.8, 170.4, 171.7, 175.4, 175.7, 178.4, 179.1.
Acetylation of 1
To the solution of 1 (200 mg) in pyridine (12 ml) was added an acetic anhydride (6 ml). The mixture was stirred at room temperature for 1 h. The solution was concentrated to dryness and the product was purified by preparative HPLC using an ODS column (SYMMETRY C18, 19 i.d. × 100 mm, Waters). The chromatography was performed with MeCN–0.3% TEAP buffer at pH 3.0 (47:53) at a flow rate of 6 ml min−1. The collected acetyl derivative of 1 (7) was extracted with ethyl acetate and the organic layer was washed with brine, and dried over anhydr. Na2SO4, and concentrated to dryness to yield 114.0 mg of 7. ESI-MS (m/z): 1241 (M+H)+. 1H NMR (500 MHz, MeOD, TMS (internal standard)): δ 0.87 (3H, d, 6.4 Hz), 0.87 (3H, d, 6.4 Hz), 0.93 (3H, d, 6.4 Hz), 0.96 (3H, d, 6.4 Hz), 0.98 (3H, d, 8.1 Hz), 1.00 (3H, d, 7.9 Hz), 1.01 (3H, s), 1.13 (3H, s), 1.15 (2H, m), 1.29 (2H, m), 1.44–1.52 (2H, overlapped), 1.52–1.59 (2H, overlapped), 1.59–1.85 (5H, overlapped), 1.80 (3H, d, 7.2 Hz), 1.91 (3H, s), 1.91 (3H, s), 1.98 (3H, s), 1.85–2.02 (2H, overlapped), 2.33 (1H, dd, 3.5, 14.1 Hz), 2.77 (1H, overlapped), 2.77 (1H, overlapped), 2.99 (1H, m), 3.08 (2H, overlapped), 3.18 (1H, dd, 3.1, 17.1 Hz), 3.22 (1H, overlapped), 3.25 (1H, overlapped), 3.49 (1H, dd, 4.6, 13.1 Hz), 3.65 (1H, dd, 4.4, 13.6 Hz), 3.77 (1H, dd, 11.7, 13.1 Hz), 4.23 (1H, dd, 6.8, 7.5 Hz), 4.44 (1H, m), 4.50 (1H, dd, 7.5, 15.4 Hz), 4.58 (1H, m), 4.74 (1H, m), 4.86 (1H, overlapped), 4.92 (1H, m), 5.06 (1H, m), 5.11 (1H, m), 5.52 (1H, q, 7.2 Hz), 7.14 (1H, m), 7.22–7.30 (4H, overlapped). 13C NMR (125 MHz, MeOD, TMS (internal standard)): δ 13.2, 20.8, 21.8, 22.6, 22.7, 22.8, 23.0, 23.1, 23.8, 23.8, 24.8, 25.7, 26.0, 27.6, 27.7, 29.1, 32.0, 33.6, 36.9, 38.1, 38.2, 39.8, 40.7, 41.2, 41.4, 42.6, 43.5, 51.2, 52.9, 53.6, 54.7, 56.2, 56.9, 59.9, 73.1, 74.0, 118.4, 128.1, 129.8, 129.8, 130.2, 130.2, 133.2, 137.7, 168.1, 170.3, 171.4, 172.0, 172.0, 172.1, 172.2, 173.4, 173.7, 174.1, 174.2, 174.3, 175.1, 175.5. One carbon signal was not observed due to overlap with the MeOD signals.
Reduction of 7 with lithium borohydride
To the solution of 7 (25 mg) in tetrahydrofuran (6.5 ml) at −78 °C was added 2.0 M lithium borohydride tetrahydrofuran solution (300 μl) followed by stirring for 1 h. The reaction was quenched by adding 10% KHSO4 solution and the product was extracted with ethyl acetate. Purification was carried out by preparative HPLC using the same ODS column described above. The chromatography was performed with MeCN–0.3% TEAP buffer at pH 3.0 (2:3) at a flow rate of 6 ml min−1. The purified reductant (8) was extracted with ethyl acetate and the organic layer was washed with brine, dried over anhydr. Na2SO4 and concentrated to dryness to yield 3.6 mg of 8. ESI-MS (m/z): 1245 (M+H)+. 1H NMR (500 MHz, MeOD, TMS (internal standard)): δ 0.87–0.92 (9H, overlapped), 0.92 (3H, d, 6.6 Hz), 0.95 (3H, d, 6.6 Hz), 0.97 (3H, d, 6.6 Hz), 1.18 (3H, s), 1.21 (2H, overlapped), 1.92 (3H, s), 1.35 (1H, m), 1.43–1.51 (3H, overlapped), 1.51–1.61 (3H, overlapped), 1.69 (1H, m), 1.72 (2H, m), 1.75 (1H, m), 1.88 (3H, d, 7.4 Hz), 1.90 (3H, s), 1.92 (3H, s), 1.92 (1H, overlapped), 1.94 (3H, s), 2.07 (1H, m), 2.29 (1H, dd, 8.9, 14.1 Hz), 2.39 (1H, dd, 3.8, 14.1 Hz), 2.53 (1H, dd, 7.2, 16.1 Hz), 2.60 (1H, dd, 6.6, 16.1 Hz), 2.97 (1H, dd, 9.1, 14.1 Hz), 3.19 (1H, m), 3.23 (2H, m), 3.43 (1H, dd, 7.0, 14.0 Hz), 3.48 (1H, dd, 5.2, 14.0 Hz), 3.59 (2H, m), 3.64 (2H, m), 3.97 (1H, m), 4.28 (1H, overlapped), 4.31 (1H, overlapped), 4.35 (1H, overlapped), 4.37 (1H, overlapped), 4.39 (1H, s), 4.43 (1H, overlapped), 4.45 (1H, overlapped), 4.64 (1H, dd, 5.4, 9.1 Hz), 5.79 (1H, q, 7.4 Hz), 7.20 (1H, m), 7.72-7.29 (4H, overlapped). 13C NMR (125 MHz, MeOD, TMS (internal standard)): δ 13.6, 21.3, 22.1, 22.7, 22.8, 22.8, 23.0, 23.1, 23.4, 23.7, 24.6, 25.8, 26.0, 26.6, 27.7, 29.2, 31.9, 36.9, 37.1, 38.2, 38.7, 40.1, 40.7, 41.1, 41.5, 41.7, 44.8, 50.1, 53.2, 53.9, 54.4, 55.3, 55.8, 56.8, 61.9, 64.2, 70.0, 72.7, 124.4, 127.9, 129.7, 129.7, 130.3, 130.3, 131.6, 138.3, 167.7, 172.2, 172.5, 172.7, 172.7, 172.7, 173.5, 174.0, 174.1, 174.4, 174.7, 174.8, 175.4.
Configuration of the constitutive amino acid
The absolute configurations of the amino acids were determined by the advanced Marfey's method.4 The acid hydrolysates of 1 (0.5 mg), 5 (0.4 mg), 6 (0.4 mg) and authentic amino acids (0.02 mg, each) were dissolved in water (10 μl). To the solution 1 M NaHCO3 (5 μl) and 1% D-FDLA/acetone solution (20 μl) (or L-FDLA/acetone solution, 20 μl) were added and incubated at 37 °C for 1.5 h. After the incubation, the reaction was terminated by the addition of 1 N HCl (5 μl). The reactant diluted with acetone was analyzed by LC-MS (LCQ spectrometer, Thermo Fisher Scientific (Waltham, MA, USA) with an ODS column (SYMMETRY C18, 4.6 i.d. × 150 mm, Waters)).
Stereochemistry of Dhb
7 (16 mg) was dissolved in a solvent composed of 0.04 M Tris–HCl buffer at pH 8.0 (9 ml). Chymotrypsin (1 mg) was dissolved in 0.001N HCl (1 ml). After pre-incubation at 37 °C for 15 min, the chymotrypsin solution (360 μl) was added to the substrate solution. The mixture was incubated at 37 °C for 20 h. After the incubation, the reaction was quenched by the addition of 50 mM aq. HCOONH4-HCOOH at pH 3.0. The digested products were isolated using a Sep-pak C18 Plus cartridge (Waters). The reaction mixture was applied to the cartridge. After washing with MeOH-5 mM aq. HCOONH4-HCOOH at pH 3.0 in the ratio of 3:7, the absorbents were eluted stepwise by using MeCN-5 mM aq. HCOONH4-HCOOH at pH 3.0 in the ratio of 3:7, 2:3 and 1:1 (5 ml, each). The peptide containing Dhb residue (9) was eluted in the 3:7 fraction. The fraction was concentrated and lyophilized to give 9 as a white powder (11.0 mg). ESI-MS (m/z): 828 (M+H)+. 1H NMR (500 MHz, DMSO-d6, TMS (internal standard)): δ 0.81 (3H, d, 6.4 Hz), 0.85 (3H, d, 5.9 Hz), 0.85 (3H, d, 5.9 Hz), 0.88 (3H, d, 6.4 Hz), 1.03 (3H, s), 1.09 (3H, s), 1.36–1.44 (1H, m), 1.46–1.63 (4H, overlapped), 1.65–1.73 (1H, m), 1.77 (3H, d, 7.3 Hz), 1.79 (3H, s), 1.79 (1H, overlapped), 1.84 (3H, s), 1.89 (1H, m), 2.36 (1H, dd, 3.1, 15.8 Hz), 2.60 (1H, dd, 10.3, 15.8 Hz), 3.16 (2H, m), 3.39 (2H, m), 3.74 (1H, br.), 4.18 (1H, m), 4.25 (1H, d, 9.2 Hz), 4.28 (1H, overlapped), 4.32 (1H, m), 4.37 (1H, m), 5.76 (1H, q, 7.3 Hz), 7.70 (1H, d, 7.9 Hz), 7.93 (1H, overlapped), 7.93 (1H, overlapped), 8.01 (1H, d, 8.1 Hz), 8.06 (1H, d, 7.2 Hz), 8.21 (1H, t, 5.2 Hz), 8.47 (1H, d, 6.6 Hz). The NH2 protons of the Dab residue, NH proton of the Dhb residue and the OH proton were observed as a broad peak. 13C NMR (125 MHz, DMSO-d6, TMS (internal standard)): δ 12.9, 21.0, 21.3, 22.4, 22.4, 22.9, 23.0, 23.9, 23.9, 25.5, 27.3, 31.7, 34.6, 40.3, 49.0, 50.8, 51.1, 52.1, 53.1, 59.8, 71.2, 119.5, 130.7, 164.5, 168.6, 169.1, 169.7, 169.8, 170.0, 171.8, 172.0, 172.6, 173.5. Three carbon signals were not observed due to overlap with the DMSO signals.
References
Wright, S. D., Ramos, R. A., Tobias, P. S., Ulevitch, R. J. & Mathison, J. C. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science 249, 1431–1433 (1990).
Kozuma, S. et al. Screeing and biological activities of pedopeptins, novel inhibitors of LPS produced by soil bacteria. J. Antibiot. 67, 237–242 (2014).
Gaillet, C., Lequart, C., Debeire, P. & Nuzillard, J. M. Band-selective HSQC and HMBC experiments using excitation sculpting and PFGSE. J. Magn. Reson. 139, 454–459 (1999).
Fujii, K., Shimoya, T., Ikai, Y., Oka, H. & Harada, K. Further application of advanced Marfey’s method for determination of absolute configuration of primary amino compound. Tetrahedron. Lett. 39, 2579–2582 (1998).
Steyn, P. L. et al. Classification of heparinolytic bacteria into a new genus, Pedobacter, comprising four species: Pedobacter heparinus comb. nov., Pedobacter piscium comb. nov., Pedobacter africanus sp. nov. and Pedobacter saltans sp. nov. proposal of the family Sphingobacteriaceae fam. nov. Int. J. Syst. Bacteriol. 48, 165–177 (1998).
Rasmussen, M. A., Madsen, S. M., Stouggard, P. & Johnsen, M. G. Flavobacterium sp. strain 4221 and Pedobacter sp. strain 4236 beta-1,3-glucanases that are active at low temperatures. Appl. Environ. Microbiol. 74, 7070–7072 (2008).
Shara, D. et al. Crystal structure of heparinase II from Pedobacter heparinus and its complex with a disaccharide product. J. Biol. Chem. 281, 15525–15535 (2006).
Margesin, R., Dieplinger, H., Hofmann, J., Sarg, B. & Lindner, H. A cold-active extracellular metalloprotease from Pedobacter cryoconitis—production and properties. Res. Microbiol. 156, 499–505 (2005).
Afonso, A., Hon, F. & Brambilla, R. Structure elucidation of Sch 20562, a glucoside cyclic dehydropeptide lactone—the major component of W-10 antifungal antibiotic. J. Antibiot. 52, 383–397 (1999).
Sano, T. & Kaya, K. Two new (E)-2-amino-2-butenoic acid (Dhb)-containing microcystins isolated from Oscillatoria agardhii. Tetrahedron 54, 463–470 (1998).
Namikoshi, M. et al. New nodularins: a general method for structure assignment. J. Org. Chem. 59, 2349–2357 (1994).
Kjaer, A. & Larsen, P. O. Amino acid studies. Part II. Structure and synthesis of albizziine (L-2-amino-3-ureidopropionic acid), an amino acid from higher plants. Acta. Chem. Scand. 13, 1565–1574 (1959).
Rao, S. L. N., Adiga, P. R. & Sarma, P. S. The isolation and characterization of β-N-oxalyl-L-α,β-diaminopropionic acid: a neurotoxin from the seeds of Lathyrus sativus. Biochemistry 3, 432–436 (1964).
Fusetani, N., Matsunaga, S., Matsumoto, H. & Takebayashi, Y. Cyclotheonamides, potent thrombin inhibitors, from a marine sponge Theonella sp. J. Am. Chem. Soc. 112, 7053–7054 (1990).
Storm, D. R., Rosenthal, K. S. & Swanson, P. E. Polymyxin and related peptide antibiotics. Annu. Rev. Biochem. 46, 723–763 (1977).
Rifkind, D. Prevention by polymyxin B of endotoxin lethality in mice. J. Bacteriol 93, 1463–1464 (1967).
Baldwin, G. et al. Effects of polymyxin B on experimental shock from Meningococcal and Escherichia coli endotoxins. J. Infect. Dis. 164, 542–549 (1991).
Sakura, N. et al. The contribution of the N-terminal structure of polymyxin B peptides to antimicrobial and lipopolysaccharide binding activity. Bull. Chem. Soc. Jpn 77, 1915–1924 (2004).
Kanazawa, K. et al. Contribution of each amino acid residue in polymyxin B3 to antimicrobial and lipopolysaccharide binding activity. Chem. Pharm. Bull. 57, 240–244 (2009).
Cowan, S. T. & Steel, K. J. in Cowan and Steel’s Manual for the Identification of Medical Bacteria 3rd edn (eds Barrow G. I., Feltham R. K. A) (Cambridge University Press: Cambridge, 1993).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Acknowledgements
We are deeply grateful to Dr Toshio Takatsu for his useful advice and Mr Yasuyuki Takamatsu for isolation of the pedopeptins.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hirota-Takahata, Y., Kozuma, S., Kuraya, N. et al. Pedopeptins, novel inhibitors of LPS: Taxonomy of producing organism, fermentation, isolation, physicochemical properties and structural elucidation. J Antibiot 67, 243–251 (2014). https://doi.org/10.1038/ja.2013.122
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2013.122
Keywords
This article is cited by
-
Nitrogen source as a modulator of the metabolic activity of Pedobacter lusitanus NL19: a transcriptomic approach
Applied Microbiology and Biotechnology (2022)
-
In-peptide amino acid racemization via inter-residue oxazoline intermediates during acidic hydrolysis
Amino Acids (2021)
-
Ogipeptins, novel inhibitors of LPS: physicochemical properties and structural elucidation
The Journal of Antibiotics (2017)
-
Identification and biological activity of ogipeptins, novel LPS inhibitors produced by marine bacterium
The Journal of Antibiotics (2017)