Abstract
Nomimicin (1), a new spirotetronate-class polyketide, was isolated from the culture broth of an actinomycete of the genus Actinomadura. Its structure was established by spectroscopic methods, and the absolute configuration was determined by a combination of NOESY experiment, J-based configuration analysis and the modified Mosher method. Nomimicin (1) showed antimicrobial activity against Micrococcus luteus, Candida albincans and Kluyveromyces fragilis.
Similar content being viewed by others
Introduction
Microbial secondary metabolites have been the most productive source of small molecules for the development of drugs.1 Almost a half of the known microbial bioactive compounds are derived from actinomycetes, and specifically Streptomyces accounts for more than 70% of the actinomycete-derived metabolites.2 Among the non-Streptomyces species, Micromonospora is the leading producer of secondary metabolites, including polyketides, peptides and glycosides, whereas Actinomadura is known as a producer of chemically and biologically unique polyketides such as antitumor enediynes,3 mannose-binding quinone glycosides with antifungal and anti-HIV activities4 and anticoccidial polyethers,5 accounting for up to 350 compounds reported as of 2005.2 During our continuing investigation on the metabolites from actinomycetes of non-Streptomyces group,6, 7 a new spirotetronate antibiotic of polyketide origin, nomimicin (1, Figure 1), was isolated from the culture extract of Actinomadura sp. TP-A0878. In this paper, we describe the isolation, structural determination and biological properties of 1.
Results and discussion
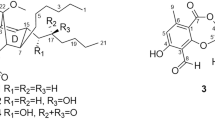
The producing strain TP-A0878 was isolated from a compost sample collected at Nomi, Ishikawa, Japan, and identified as a member of Actinomadura on the basis of 16S rRNA gene sequence. This strain was cultured in A-3M medium at 30 °C for 6 days, and the whole culture broth was extracted with 1-butanol. The extract was consecutively fractionated by normal- and reversed-phase column chromatographies, followed by HPLC purification on a C18 column, to yield nomimicin (1) as an optically active, amorphous solid ([α]D –94, CHCl3). Compound 1 gave a pseudomolecular ion at m/z 519.2726 [M+Na]+ in a HR-ESI time-of-flight (TOF) MS measurement, suggesting a molecular formula of C30H40O6, which was subsequently corroborated by the NMR data. The IR spectrum indicated the presence of hydroxyl (3397, cm−1) and carbonyl (1749, cm−1) functionalities. Analysis of 1H and 13C NMR, and HSQC data established three oxygenated quaternary sp2 carbons, seven sp2 carbons (five are proton-bearing), three quaternary sp3 carbons (one is oxygen-bearing), seven sp3 methines (two are oxygen-bearing), five sp3 methylenes and five methyl groups (Table 1). The UV spectrum of 1 was closely similar to that of maklamicin7 (Figure 1), a spirotetronate polyketide that we recently discovered from a Micromonospora strain, suggesting the presence of a tetronic acid functionality in this molecule. This was supported by the 13C chemical shifts of the carbons C-1 (δC 167.0), C-2 (δC 107.2), C-3 (δC 200.3) and C-24 (δC 204.5) that closely matched those reported for this structure in maklamicin. As three double bonds and a tetronic acid unit that contains three double bonds and one ring accounted 7 of the 11 double-bond equivalents, 1 must possess four more rings to satisfy the molecular formula.
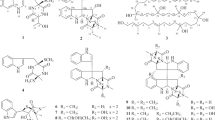
Interpretation of the COSY and HSQC spectra provided six fragments (indicated by bold lines, Figure 2), and the linkage among these fragments was established by analysis of HMBC correlations. A secondary methyl fragment C-8/C-26 and two large fragments C-7/C-6/C-5/C-10/C-9 and C-11–C-17 were joined to form an oxygenated cyclohexane substructure, based on HMBC correlations from H3-26 to C-7 and C-9, H-11 to C-5 and C-9, and H-9 to C-11. Furthermore, a series of long-range correlations from a singlet methyl proton resonance H3-25 to C-3, C-4, C-5 and C-13 fused an additional ring to the cyclohexane ring. Accordingly, a Δ1-octalin framework with an olefinic extension (C-14 to C-17) and a carbonyl functionality from the tetronic acid unit (C-3) were established to be present in the bottom half of the molecule. The E-geometry of the olefinic extension was deduced from a large vicinal coupling constant between H-15 and H-16 (15.3 Hz). The remaining fragments C-21/C-22, C-29/C-30 and C-19/C-20/C-28 were established by COSY correlations, and were assembled by HMBC correlations from H3-28 to C-19, C-20 and C-21, H-30 to C-21, and H-29 to C-20 and C-22 into the upper half of the cyclohexene unit in the top half of the molecule. Finally, a set of HMBC correlations from H3-27 to C-17, C-18, C-19 and C-23 assigned an oxygenated spirocarbon (C-23) and a quaternary carbon C-18 to be placed in the lower half of the cyclohexene ring, and at the same time, as a juncture to the tetronic acid moiety and to the terminus of the octalin extension, respectively. The spiro connection between the cyclohexene and tetronic acid units was also supported by an HMBC correlation from H2-22 to C-24, and thus a characteristic pentacycle-condensed spirotetronate structure was assigned for 1 (Figure 2).
The relative configuration was elucidated by NOESY experiments and J-based configuration analysis (Figures 3 and 4). NOEs between H-5, H-7 and H-9, and large scalar couplings (3JHH>10 Hz) shown by H-5/H-6α, H-6α/H-7 and H-5/H-10 provided the 1,3-diaxial relationships between H-5, H-7 and H-9, and a trans ring fusion in the octalin unit. The axial orientation of the methyl group at C-8 was confirmed by an NOE between H3-26 and H-10. NOEs between H-10 and H3-25, and between H3-25 and H-13 placed the H3-25 methyl and H-13 on the same side of the octalin ring, with respect to the axial H-10 proton. A series of NOESY correlations for H-13/H-15, H-15/H-17b, H-17a/H-19, H-14b/H-16 and H-16/H3-27, established a zigzag conformation of the C-13 to C-18 chain, and the configuration at C-18 relative to C-13. Furthermore, NOEs between H-21 and H-22β, and between H-22β and H3-27 established that these protons were located on the same side of the cyclohexene ring (Figure 3). Finally, the configuration at the spirocarbon C-23 was determined using the J-based configuration analysis.8 Heteronuclear long-range coupling constants 2JCH and 3JCH were determined by J-resolved HMBC experiments.9, 10 J-Resolved HMBC spectra were measured on a Varian INOVA-500 spectrometer at 20 °C using a microtube (Shigemi Inc., Tokyo, Japan) in CDCl3 (10 mg of 1 in 0.25 ml). In the J-resolved HMBC spectra, 2JCH and 3JCH values are obtained as absolute values. The small coupling constant between H-22β and C-23 (2JCH<2 Hz), and the large coupling constant between H-22α and C-23 (2JCH=5 Hz) suggested the anti-relationship of H-22β and the oxygen atom at C-23 (Figure 4).
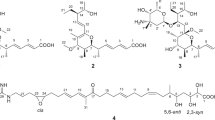
The absolute stereochemistry was determined by applying the modified Mosher method11 to the secondary hydroxyl groups at C-7 and C-9, both of which were in equatorial orientation. After methylation of the enolic hydroxyl group at C-24 with TMSCHN2 in CHCl3/MeOH, the methylated product 2 was reacted with (S)- and (R)-MTPA (α-methoxy-α-trifluoromethylphenylacetyl) chloride, yielding bis-(R)- and (S)-MTPA esters (3 and 4), respectively. Calculation of the ΔδH(S-R) values identified H-11 to H2-17 to have positive signs, whereas H2-6 and H3-25 negative signs (Figure 5). These data allowed assignment of both the absolute configurations at C-7 and C-9 as R. Although signs were mixed for the protons flanked by the MTPA ester groups, the sign distribution pattern is in good accordance with that reported for bisMTPA derivatives of cyclohexane-1,3-diols.12 The deduced absolute configuration was identical with that known for this antibiotic family.13, 14
Spirotetronates represented by kijanimicin15 feature a trans-Δ1-octalin unit and a tetronic acid moiety spiro-linked with a cyclohexene ring. To date, over 50 related metabolites have been discovered from actinomycetes. Their structural variations are largely derived from the glycosylation pattern and the length of the carbon chain connecting the octalin and the cyclohexene units. Maklamicin is the first example to have the shortest four-carbon linker, and nomimicin (1) is a new member of this class.
Nomimicin (1) showed antimicrobial activities against Micrococcus luteus, Candida albicans and Kluyveromyces fragilis with MIC values of 6.3, 12.5 and 12.5 μg ml−1, respectively, whereas it was inactive against Escherichia coli. In contrast, maklamicin is not active against these yeasts, suggesting the effect of the hydroxylation pattern on antifungal activities. Compound 1 displayed weak cytotoxicity against human cervical cancer cells HeLa and human breast cancer cells MCF7, with IC50 values of 74 and 59 μM, respectively.
Experimental procedures
General experimental procedures
Optical rotations were measured using a JASCO DIP-3000 polarimeter (JASCO Corporation, Tokyo, Japan). UV spectrum was recorded on a Hitachi U-3210 spectrophotometer (Hitachi, Tokyo, Japan). IR spectrum was measured on a Perkin Elmer Spectrum 100 (Perkin-Elmer, Fremont, CA, USA). 13C NMR spectra were measured on a Bruker AVANCE 400 spectrometer, and 1H and 2D NMR spectra were obtained on a Bruker AVANCE 500 spectrometer (Bruker, Rheinstetten, Germany). Chemical shifts were referenced to residual solvent signals (δH 7.26, δC 77.0). J-Resolved HMBC experiments were performed on a Varian INOVA-500 spectrometer (Varian Inc., Palo Alto, CA, USA). HR-ESITOFMS were recorded on a Bruker microTOF focus. Silica Gel 60 (Kanto Chemical Co., Inc., Tokyo, Japan; 63-210 mesh) and Silica Gel 60-C18 (Nacalai Tesque, Kyoto, Japan; 250-350 mesh) were used for silica gel and octa decyl silica (ODS) column chromatographies, respectively. HPLC separation was performed using an XTerra RP18 (Waters Corporation, Milford, MA, USA; 7 μm, 19 × 300 mm) with a photodiode array detector.
Producing microorganism
Strain TP-A0878 was isolated from a compost sample collected at Nomi, Ishikawa, Japan, in 2007. The strain was identified as a member of the genus Actinomadura on the basis of 100% identity of 16S rRNA gene sequence (1462 nucleotides; DDBJ accession number AB488798) with Actinomadura sp. TFS 455 (accession number EF2120220).
Fermentation
Strain TP-A0878 cultured on a Bn-2 slant (soluble starch 0.5%, glucose 0.5%, meat extract (Kyokuto Pharmaceutical Industrial Co., Ltd., Tokyo, Japan) 0.1%, yeast extract (Difco Laboratories, Becton, Dickinson and Company, Sparks, MD, USA) 0.1%, NZ-case (Wako Chemicals USA, Inc., Richmond, VA, USA) 0.2%, NaCl 0.2%, CaCO3 0.1%, agar 1.5%) was inoculated into 500-ml K-1 flasks, each containing 100 ml of V-22 seed medium consisting of soluble starch 1%, glucose 0.5%, NZ-case (Wako Chemicals USA, Inc.) 0.3%, yeast extract (Difco Laboratories) 0.2%, tryptone (Difco Laboratories) 0.5%, K2HPO4 0.1%, MgSO4·7 H2O 0.05% and CaCO3 0.3% (pH 7.0). The flasks were placed on a rotary shaker (200 r.p.m.) at 30 °C for 4 days. The seed culture (3 ml) was transferred into 500-ml K-1 flasks, each containing 100 ml of A-3M production medium consisting of glucose 0.5%, glycerol 2%, soluble starch 2%, Pharmamedia (Traders Protein, Lubbock, TX, USA) 1.5%, yeast extract 0.3% and Diaion HP-20 (Mitsubishi Chemical Co., Tokyo, Japan) 1%. The pH of the medium was adjusted to 7.0 before sterilization. The inoculated flasks were placed on a rotary shaker (200 r.p.m.) at 30 °C for 6 days.
Extraction and isolation
At the end of the fermentation period, 100 ml of 1-butanol were added to each flask and they were allowed to shake for 1 h. The mixture was centrifuged at 6000, r.p.m. for 10 min, and the organic layer was separated from the aqueous layer containing the mycelium. Evaporation of the solvent gave 3.0 g of the crude extract from 2-l culture. This was subjected to silica gel column chromatography with a stepwise gradient of CHCl3/MeOH (1:0, 20:1, 10:1, 4:1, 2:1, 1:1 and 0:1 v/v). Fractions 5 (2:1) and 6 (1:1) were concentrated to provide 0.47 g of a brown solid, which was further purified by reversed-phase ODS column chromatography with a gradient of MeCN/0.15% KH2PO4 buffer (pH 3.5; 2:8, 3:7, 4:6, 5:5, 6:4, 7:3 and 8:2 v/v). Fraction 6 (7:3) was evaporated to an aqueous solution, which was extracted with EtOAc. The organic layer was dried over anhydrous Na2SO4, filtered and concentrated to give a pale yellow solid (90 mg). Final purification was achieved by repeated preparative HPLC with MeCN/0.1% HCO2H (MeCN concentration: 50% for 0–8 min; 50–80% for 8–42 min; 14 ml min−1; UV detection at 254 nm), followed by evaporation and extraction with EtOAc, to yield nomimicin (1, 19 mg, tR 22.5 min).
Nomimicin (1)
Colorless amorphous solid; [α]23D –94 (c 0.10, CHCl3); UV (MeOH) λmax (log ɛ) 250 (3.97), 293 (3.89) nm; IR (attenuated total reflection) νmax 3397, 1749, cm−1; 1H and 13C NMR data, see Table 1; HR-ESITOFMS [M+Na]+ 519.2726 (calcd for C30H40O6Na, 519.2717).
Methylation of 1 to yield 2
24-O-Methyl ether of 1 (2): To a solution of 1 (5.0 mg, 0.010 mmol) in CHCl3/MeOH (0.25 ml each) was added a solution of TMSCHN2 in Et2O (2.0 M, 0.25 ml, 0.50 mmol) at room temperature. After stirring for 15 min, the reaction mixture was concentrated to dryness. The residue was purified on a silica-gel column chromatography (hexane/EtOAc=20:1–1:1) to give 2 (3.5 mg) in 60% yield: colorless amorphous solid; [α]25D –52 (c 0.50, CHCl3); 1H NMR (500 MHz, CDCl3) δ 0.91 (3 H, t, J=7.6 Hz, H-30), 0.96 (3 H, d, J=7.0 Hz, H-26), 1.04 (3 H, s, H-27), 1.20 (1 H, ddd, J=11.9, 11.9, 11.7 Hz, H-6α), 1.72 (3 H, s, H-28), 1.59 (1 H, m, H-6β), 1.53 (3 H, s, H-25), 1.60 (1 H, m, H-5), 1.62 (1 H, m, H-29a), 1.66 (1 H, m, H-29b), 1.66 (1 H, m, H-22α), 1.84 (1 H, m, H-17a), 1.85 (2 H, m, H-14), 1.88 (1 H, m, H-21), 1.91 (1 H, m, H-10), 2.18 (1 H, dd, J=14.6, 7.1 Hz, H-22β), 2.38 (1 H, m, H-8), 2.40 (1 H, m, H-17b), 2.87 (1 H, br.dd, J=5.0, 5.0 Hz, H-13), 3.43 (1 H, dd, J=10.7, 4.7 Hz, H-9), 3.95 (1 H, ddd, J=11.7, 4.3, 4.3 Hz, H-7), 4.07 (3 H, s, 24-OCH3), 4.86 (1 H, s, H-19), 5.02 (1 H, dd, J=15.1, 11.4 Hz, H-16), 5.30 (1 H, dddd, J=15.1, 10.7, 2.1, 2.1 Hz, H-15), 5.57 (1 H, ddd, J=10.0, 5.0, 2.4 Hz, H-12), 5.81 (1 H, d, J=10.0 Hz, H-11); 13C NMR (100 MHz, CDCl3) δ 5.4 (C-26), 13.0 (C-30), 15.5 (C-25), 22.2 (C-28), 23.7 (C-27), 24.8 (C-29), 29.6 (C-6), 29.7 (C-22), 35.4 (C-5), 35.9 (C-14), 36.4 (C-10), 37.6 (C-13), 39.2 (C-21), 39.3 (C-18), 41.8 (C-8), 43.6 (C-17), 51.6 (C-4), 65.8 (24-OCH3), 71.0 (C-7), 74.1 (C-9), 86.0 (C-23), 111.9 (C-2), 123.4 (C-11), 126.9 (C-16), 129.7 (C-19), 131.4 (C-15), 131.7 (C-12), 133.7 (C-20), 168.2 (C-1), 193.7 (C-24), 196.6 (C-3); HR-ESITOFMS [M+Na]+ 533.2865 (calcd for C31H42O6Na, 533.2874).
Bis-(R)-MTPA ester of 2 (3)
(S)-MTPA chloride (5.0 μl, 27 μmol) was added to a solution of 2 (1.7 mg, 3.3 μmol) in dry pyridine (1 ml) at room temperature. After 18 h, the reaction mixture was concentrated to dryness and purified by silica-gel column chromatography (n-hexane/EtOAc=1:0-1:1), to yield 3 (1.6 mg, 42%): 1H NMR (500 MHz, CDCl3) δ 0.87 (3 H, d, J=7.0 Hz, H-26), 0.91 (3 H, t, J=7.4 Hz, H-30), 1.04 (3 H, s, H-27), 1.34 (1 H, ddd, J=12.0, 12.0, 12.0 Hz, H-6α), 1.73 (3 H, s, H-28), 1.79 (1 H, m, H-14), 1.82 (1 H, m, H-5), 1.82 (1 H, m, H-6β), 1.85 (1 H, m, H-14), 1.85 (1 H, m, H-17a), 2.20 (1 H, m, H-10), 2.40 (dd, J=14.3, 11.5 Hz, H-17b), 2.78 (1 H, m, H-8), 2.83 (1 H, dd, J=5.0, 5.0 Hz, H-13), 4.87 (1 H, s, H-19), 4.91 (1 H, dd, J=11.5, 4.7 Hz, H-9), 5.05 (1 H, dd, J=14.8, 11.5 Hz, H-16), 5.10 (1 H, d, J=10.0 Hz, H-11), 5.28 (1 H, m, H-15), 5.30 (1 H, m, H-7), 5.48 (1 H, ddd, J=10.0, 5.0, 2.4 Hz, H-12); HR-ESITOFMS m/z 965.3652 [M+Na]+ (calcd for C51H56F6O10Na, 965.3670).
Bis-(S)-MTPA ester of 2 (4)
In the same manner as described for 3, 4 (1.3 mg) was prepared from 1.7 mg of 2: 1H NMR (500 MHz, CDCl3) δ 0.73 (3 H, d, J=7.0 Hz, H-26), 0.91 (3 H, t, J=7.4 Hz, H-30), 1.05 (3 H, s, H-27), 1.25 (1 H, ddd, J=11.7, 11.7, 11.7 Hz, H-6α), 1.72 (1 H, m, H-6β), 1.73 (3 H, s, H-28), 1.80 (1 H, m, H-14), 1.82 (1 H, m, H-5), 1.85 (1 H, m, H-14), 1.86 (1 H, m, H-17a), 2.20 (1 H, m, H-10), 2.41 (dd, J=14.0, 11.7 Hz, H-17b), 2.79 (1 H, m, H-8), 2.85 (1 H, dd, J=5.0, 5.0 Hz, H-13), 4.86 (1 H, dd, J=11.6, 4.7 Hz, H-9), 4.88 (1 H, s, H-19), 5.06 (1 H, dd, J=14.9, 11.7 Hz, H-16), 5.28 (1 H, m, H-7), 5.29 (1 H, m, H-15), 5.32 (1 H, d, J=10.0 Hz, H-11), 5.56 (1 H, ddd, J=10.0, 5.0, 2.4 Hz, H-12); HR-ESITOFMS m/z 965.3679 [M+Na]+ (calcd for C51H56F6O10Na, 965.3670).
Biological assays
Antimicrobial assay16 and cytotoxic assay17 were carried out according to the procedures previously described.
References
Butler, M. S. Natural products to drugs: natural product-derived compounds in clinical trials. Nat. Prod. Rep. 25, 475–516 (2008).
Bérdy, J. Bioactive microbial metabolites. J. Antibiot. 58, 1–26 (2005).
Smith, A. L. & Nicolaou, K. C. The enediyne antibiotics. J. Med. Chem. 39, 2103–2117 (1996).
Walsh, T. J. & Giri, N. Pradimicins: a novel class of broad-spectrum antifungal compounds. Eur. J. Clin. Microbiol. Infect. Dis. 16, 93–97 (1997).
Dirlam, J. P. et al. CP-84,657, a potent polyether anticoccidial related to portmicin and produced by Actinomadura sp. J. Antibiot. 43, 668–679 (1990).
Igarashi, Y. et al. Brartemicin, an inhibitor of tumor cell invasion from the actinomycete Nonomuraea sp. J. Nat. Prod 72, 980–982 (2009).
Igarashi, Y. et al. Maklamicin, an antibacterial polyketide from an endophytic Micromonospora sp. J. Nat. Prod. 74, 670–674 (2011).
Matsumori, N., Kaneno, D., Murata, M., Nakamura, H. & Tachibana, K. Stereochemical determination of acyclic structures based on carbon–proton spin-coupling constants. A method of configuration analysis for natural products. J. Org. Chem. 64, 866–876 (1999).
Furihata, K. & Seto, H. J-Resolved HMBC, a new NMR technique for measuring heteronuclear long-range coupling constants. Tetrahedron Lett. 40, 6271–6275 (1999).
Furihata, K., Tashiro, M. & Seto, H. Selective J-resolved HMBC, an efficient method for measuring heteronuclear long-range coupling constants. Magn. Reson. Chem. 47, 814–818 (2009).
Ohtani, I., Kusumi, T., Kashman, Y. & Kakisawa, H. High-field FT NMR application of Mosher’s method. The absolute configurations of marine terpenoids. J. Am. Chem. Soc. 113, 4092–4096 (1991).
Konno, K., Fujishima, T., Liu, Z. & Takayama, H. Determination of absolute configuration of 1,3-diols by the modified Mosher’s method using their di-MTPA esters. Chirality 13, 72–80 (2002).
Momose, I. et al. Decatromicins A and B, new antibiotics produced by Actinomadura sp. MK73-NF4. II. Structure determination. J. Antibiot. 52, 787–796 (1999).
Park, H-R., Chijiwa, S., Furihata, K., Hayakawa, Y. & Shin-ya, K. Relative and absolute configuration of versipelostatin, a down-regulator of molecular chaperone GRP78 expression. Org. Lett. 9, 1457–1460 (2007).
Mallams, A. K., Puar, M. S., Rossman, R. R., McPhail, A. T. & Macfarlane, R. D. Kijanimicin. 2. Structure and absolute stereochemistry of kijanimicin. J. Am. Chem. Soc. 103, 3940–3943 (1981).
Igarashi, Y. et al. Abyssomicin I, a modified polycyclic polyketide from Streptomyces sp. CHI39. J. Nat. Prod. 73, 1943–1946 (2010).
Fukuda, T. et al. Marianins A and B, prenylated phenylpropanoids from Mariannaea camptospora. J. Nat. Prod. 74, 1327–1330 (2011).
Acknowledgements
We acknowledge Dr T Okuda and Ms Y Sudoh at Tamagawa University for assistance with antimicrobial assay.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Igarashi, Y., Iida, T., Oku, N. et al. Nomimicin, a new spirotetronate-class polyketide from an actinomycete of the genus Actinomadura. J Antibiot 65, 355–359 (2012). https://doi.org/10.1038/ja.2012.30
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2012.30
Keywords
This article is cited by
-
Development of a drug discovery approach from microbes with a special focus on isolation sources and taxonomy
The Journal of Antibiotics (2023)
-
Promising bioactive compounds from the marine environment and their potential effects on various diseases
Journal of Genetic Engineering and Biotechnology (2022)
-
Actinomadura decatromicini sp. nov., isolated from mountain soil in Thailand
The Journal of Antibiotics (2021)
-
Structure elucidation and in silico docking studies of a novel furopyrimidine antibiotics synthesized by endolithic bacterium Actinomadura sp. AL2
World Journal of Microbiology and Biotechnology (2017)
-
Three antimycobacterial metabolites identified from a marine-derived Streptomyces sp. MS100061
Applied Microbiology and Biotechnology (2013)