Abstract
A new ryanodine-binding inhibitor, verticilide, was isolated from the cultured broth of a fungus, Verticillium sp. FKI-1033. It is a 24-membered ring cyclic depsipeptide, its structure being elucidated as cyclo[(2R)-2-hydroxyheptanoyl-N-methyl- L-alanyl]4. Verticilide inhibited ryanodine binding to ryanodine receptors in the cockroach at an IC50 value of 4.2 μM, whereas inhibition against mouse ryanodine receptors was weak (IC50=53.9 μM).
Similar content being viewed by others
Introduction
In the course of screening for insecticidal compounds, we have isolated some new antibiotics from microbial metabolites.1 Recently, our attention turned to ryanodine receptors as a potential target for new insecticides.
Ryanodine was first isolated as an insecticide from the stem and roots of the plant Ryania speciosa. It was shown to bind Ca2+ channel of the sarcoplasmic reticulum, leading to the discovery of ryanodine receptors. A ryanodine receptor (RyR) is a Ca2+ release channel found in the intracellular sarcoplasmic or endoplasmic reticulum of a variety of cells.2, 3 There are three different isoforms (RyR1, RyR2 and RyR3) found in mammals. RyR1 is abundant in the skeletal muscle, RyR2 is abundant in the cardiac muscle and RyR3 is found in the brain and also at lower levels in a variety of tissues. RyRs control intracellular Ca2+ release in skeletal and cardiac muscles, activating muscle contraction during excitation–contraction (EC) coupling. In skeletal muscle, EC coupling occurs through voltage-gated Ca2+ release, controlled by direct protein–protein interactions between L-type Ca2+ channel at the plasma membrane and RyR1. Conversely, RyR2 and RyR3 are activated by elevated Ca2+ concentrations through the L-type Ca2+ channel, which is called Ca2+-induced Ca2+ release.
Dantrolene is a drug used for malignant hyperthermia, which is a pharmacogenetic disorder triggered by halogenated anesthetics, and is typically inhaled during surgery. Dantrolene is believed to inhibit RyR1 and RyR3 but not RyR2.4 The RyRs are potential therapeutic targets for diverse human disorders of cardiac and skeletal muscles and of the central nervous system.
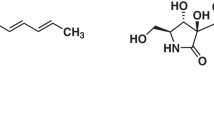
In contrast to mammalian receptors, insects have a single but distinct RyR.5 Therefore, insect RyR is a potential target for new insecticides.6 The molecular target of some new synthetic insecticides, flubendiamide and anthranilic diamide, was shown to be RyR.7, 8 Consequently, we screened for ryanodine-binding inhibitors specific to insect RyR from microbial origins and isolated a new compound, verticilide (1, Figure 1), produced by a cultured broth of Verticillium sp. FKI-1033.9
We have already reported the stereochemical study and the total synthesis of 1.10 In this report, we describe the taxonomy of the producing strain, the fermentation, isolation and the planar structure elucidation with detailed NMR assignments of 1 and its ryanodine-binding inhibitory activity compared with the activity of the structure-related compounds. The other biological activities of 1 are also described.
Results and discussion
Taxonomy of the producing organism
Strain FKI-1033 was originally isolated from a soil sample collected in Kagoshima Prefecture, Japan. Colonies of strain FKI-1033 grown on potato dextrose agar were 42–44 mm in diameter after 7 days at 25 °C, floccose to velvety and white in color. The reverse side was white to pale-yellowish brown. Microscopic observation of colonies grown on malt extract agar identified hyphae with septa, with conidiophores born from the aerial mycelia, with occasional branching (Figure 2). The phialide was observed directly from aerial hyphae, or alone or two to four verticillations in the middle part or top of conidiophores. The phialide was 25–80 μm with slight swelling at the base (2.0–2.8 μm) and narrowing into a conical shape at the top. Conidia were suglobose to oval (2.5–4.0 × 2.0–3.0 μm) and formed a viscous conidial mass at the top of the phialide.
From the above characteristics, the microorganism was considered to belong to the genus Verticillium Nees11 and was named Verticillium sp. FKI-1033. The strain was deposited at the International Patent Organism Depositary, National Institute of Advanced Industrial Science and Technology, Tsukuba, Japan, as FERM BP-8219.
Fermentation
A stock culture of strain FKI-1033 was inoculated into a 500-ml Erlenmeyer flask containing 100 ml of a seed medium and incubated on a rotary shaker at 27 °C for 3 days. The seed broth was diluted to five times with sterilized water and 10 ml of the diluted broth was transferred into each of twenty-four 1-l Roux flasks containing 240 g of a production medium. Fermentation was carried out statically at 27 °C for 13 days.
Isolation
After 300 ml of methanol–H2O (2:1) was added to each flask, they were vigorously shaken and then kept for 3 h. The extract solutions in each flask were combined, the methanol in the solution was removed by evaporation and the remaining water solution was partitioned with ethyl acetate twice, the organic layer finally being concentrated in vacuo to afford a brown oil (2.53 g). This was applied to a silica gel column (ø 2.8 × 14 cm, Silica gel 60, Merck 107734, Merck KGaA, Darmstadt, Germany) and washed with hexane–ethyl acetate (4:3). Active fractions eluted with hexane–ethyl acetate (2:3) were concentrated to yield a brown oil (183 mg), which was applied to a Sephadex LH-20 column (GE Healthcare UK Ltd., Little Chalfont, UK) (ø 2.7 × 92 cm) and eluted with methanol to afford a colorless oil of 1 (86.5 mg).
Structure elucidation
The physico-chemical properties of 1 are summarized in Table 1. The molecular formula of 1 was established as C44H76N4O12 using HR-FAB-MS. The IR absorbances observed at 1743 and 1662 cm–1 suggested the presence of ester carbonyl groups and amide carbonyl groups, respectively. The 1H and 13C NMR of 1 showed very complicated spectra. Their chemical shifts are shown in Table 2. The signals of δH 4.5–5.6 and δc 51–55 suggested α-carbons of amino acids and the signals of δc 169–172 and the IR absorbances (see above) suggested ester carbonyl and amide carbonyl carbons.
The initial structure elucidation of 1 was thus carried out using its acid hydrolysis, which afforded N-methyl-L-alanine (2) and (2R)-2-hydroxyheptanoic acid (3) as reported previously.10 Mild alkaline hydrolysis (0.24 M sodium hydroxide for 24 h) of 1 afforded 4, which was elucidated as 2-hydroxyheptanoyl-N-methylalanine using spectroscopic data.
The molecular formula of 1 suggested that it is a cyclic tetramer of 4, and the 1H and 13C NMR spectra of 1 are similar to those of 4. However, >50 carbon signals were observed in the 13C NMR of 1, which suggests that it exists in two conformers. Detailed analysis, using COSY and heteronuclear multiple bond correlation experiments of 1, revealed that the ratio of conformer A (asymmetric conformer) and conformer B (symmetric conformer) is 3:4 (Table 2 and Figure 3). Similar conformer mixtures have been reported for symmetric 24-membered ring depsipeptides, bassianolide and PF1022A.12, 13 NMR spectra of PF1022A are known to be simplified by addition of potassium thiocyanate (M Ohyama, personal communication). When the NMR spectra were observed in CD3OD with potassium thiocyanate (30 mM of 1 and 300 mM of potassium thiocyanate), the spectra showed only one symmetric conformer (Table 2). Thus, the structure of 1 was elucidated as cyclo[(2R)-2-hydroxyheptanoyl-N-methyl-L-alanyl]4 (Figure 1). This was confirmed using total synthesis.10
Although microorganisms produce various cyclic depsipeptides, there are few reports of natural 24-membered ring depsipeptides that contain alternate arrangements of amino and hydroxy acids. The antifungal amidomycin, cyclo[(2R)-2-hydroxy-3-methylbutanoyl-D-valyl]4, and cytotoxic montanastatin, cyclo[(2R)-2-hydroxy-3-methylbutanoyl-D-valyl-(2S)-2-hydroxypropanoyl-L-valyl]2, are produced by Streptomyces spp.14, 15 As for fungi, bassianolide, cyclo[(2R)-2-hydroxy-3-methylbutanoyl-N-methyl-L-leucyl]4, is produced by Beauveria bassiana and Verticillium lecanii,12, 16 whereas PF1022 group compounds (types A–H) are produced by Rosellinia sp.13, 17, 18, 19, 20 The structure of PF1022A is cyclo[(2R)-2-hydroxypropanoyl-N-methyl-L-leucyl-3-phenyl-(2R)-2-hydroxypropanoyl-N-methyl-L-leucyl]2. Therefore, fungal compounds have R-form hydroxy acids and L-form amino acids, which is in accordance with the stereochemistry of 1. Bassianolide has an insecticidal activity, and is reported to inhibit the tonic component of contraction induced by acetylcholine in the guinea pig.21 Emodepside, a semisynthetic derivative of PF1022A, with a morpholine ring at each of the two phenyl rings in the para position, is now used as a nematocide in the veterinary field. The target of emodepside and PF1022A is suggested to be depsiphilin, which is similar to mammalian latrophilin, a presynaptic G-protein-coupled receptor.22
Biological activities
Compound 1 showed ryanodine-binding inhibition against cockroach RyR with an IC50 value of 4.2 μM (Table 3). The IC50 value of ryanodine-binding inhibition against mouse leg muscle RyR was 53.9 μM, indicating that it was more than 10 times weaker when compared with the cockroach receptor. Ryanodine did not show high selectivity between cockroach and mouse receptors.
Compound 1 belongs to a group of cyclic depsipeptides having alternately combined amino acids and hydroxy acids. However, 1 is the first compound that has small side-chain amino acids and very long, straight side-chain hydroxy acids, and hence its biological activity is interesting. We consequently studied the ryanodine-binding inhibitory activity of some cyclic depsipeptides—bassianolide, PF1022A, beauvericin, and enniatins A, A1, B, B1, D and F (Figure 1). Beauvericin and the enniatins are 18-membered ring depsipeptides. All depsipeptides showed no ryanodine-binding inhibition against cockroach RyR at 100 μg ml–1. Therefore, it is likely that the unique side-chain structure in 1 could contribute to its ryanodine-binding inhibition.
Insecticidal and nematocidal activities of 1 were studied by a microplate assay using brine shrimp, Artemia salina, and the free-living nematode Caenorhabditis elegans. The minimum growth inhibitory concentrations of 1 against C. elegans and A. salina were both 20 μg ml–1. Compound 1 did not inhibit the growth of P388 cells at 100 μg ml–1 and it did not show any antimicrobial activity against some Gram-positive and Gram-negative bacteria, fungi and yeasts at 10 μg disk–1 (paper disk method).
Experimental section
General
NMR spectra were recorded on a Varian Inova 600 spectrometer (Varian Inc., Palo Alto, CA, USA) (2–3JCH=8 Hz in heteronuclear multiple bond correlation). Chemical shifts are shown in δ values (p.p.m.) relative to chloroform at 7.26 p.p.m. for 1H NMR and at 77.0 p.p.m. for 13C NMR. MS was conducted on a JEOL JMS-AX505 HA spectrometer (JEOL Ltd., Akishima, Japan). The UV and IR spectra were measured with a Shimadzu UV-240 spectrophotometer (Shimadzu Corporation, Kyoto, Japan) and a Horiba FT-210 Fourier (Horiba Ltd., Kyoto, Japan) transform infrared spectrometer, respectively. Optical rotations were recorded on a JASCO model DIP-181 polarimeter (JASCO Corporation, Hachioji, Japan).
Taxonomy of the producing organism
Morphological observations of the verticilide-producing strain were carried out using an Olympus Vanox-S AH-2 microscope (Olympus Corporation, Tokyo, Japan).
Media
The seed medium consisted of glucose 2.0%, Polypepton (Nihon Pharmaceutical Co. Ltd, Tokyo, Japan) 0.5%, yeast extract (Oriental Yeast Co. Ltd., Tokyo, Japan) 0.2%, KH2PO4 0.1%, MgSO4·7H2O 0.05% and agar 0.1%, pH 6.0. The production was carried out in 1-l Roux flasks, each containing Italian rice 150 g, FeSO4·7H2O 0.9 mg, MnCl2·4H2O 0.9 mg, ZnSO4·7H2O 0.9 mg, CuSO4·5H2O 0.9 mg, CoCl2·6H2O 0.9 mg and tap water (90 ml).
Acid hydrolysis of 1
Compound 1 (50.0 mg) was hydrolyzed with 6 M hydrochloric acid (HCl; 2.5 ml) at 110 °C for 24 h in a sealed tube. The reaction mixture was concentrated under reduced pressure. The residue was applied to a Dowex 50W × 8 column (H+ form, ø 1.7 × 9.8 cm) and eluted with water and 1 M ammonia. The water eluate was concentrated to dryness to yield a white powder of 3 (20.2 mg). The ammonia eluate was concentrated to yield a colorless oil of 2 (12.4 mg).
N-Methyl-L-alanine (2)
[α]D27 +6.44 (c 0.56, 6 M HCl); FAB-MS (m/z) 104 (M+H)+; 1H NMR (600 MHz, D2O) δ 3.45 (1H, q, Jα,β=6.6 Hz, α-CH), 2.53 (3H, s, NCH3), 1.31 (3H, d, Jα,β=6.6 Hz, β-CH3); 13C NMR (150 MHz, D2O) δ 175.1 (C=O), 59.1 (α-CH), 31.2 (NCH3), 14.7 (β-CH3). The stereochemistry was found to be the L-form by chiral HPTLC (CHIR, Merck 114101; developed by 0.05 M KH2PO4–methanol–CH3CN (5:5:1)).
(2R)-2-Hydroxyheptanoic acid (3)
[α]D25 –15.46 (c 1.0, CHCl3); HR-FAB-MS (m/z) found 145.0866, calcd 145.0865 [(M–H)–, C7H13O3]; 1H NMR (600 MHz, D2O) δ 4.12 (1H, m, α-CH), 1.65 (1H, m, β-CH2), 1.55 (1H, m, β-CH2), 1.24 (2H, m, γ-CH2), 1.15 (4H, m, δ-CH2 and ɛ-CH2), 0.71 (3H, m, ζ-CH3); 13C NMR (150 MHz, D2O) δ 178.6 (C=O), 70.6 (α-CH), 33.5 (β-CH2), 30.9 (δ-CH2), 24.0 (γ-CH2), 22.1 (ɛ-CH2), 13.6 (ζ-CH3).
Alkaline hydrolysis of 1
After addition of 1 M sodium hydroxide (1.6 ml) to a solution of 1 (50.0 mg) in methanol (5.0 ml), the mixture was stirred for 24 h at room temperature. The reaction mixture was neutralized with 1 M HCl and concentrated under reduced pressure. The concentrate was purified using HPLC on a Senshu Pak C18 column (ø 20 × 250 cm, Senshu Scientific Co. Ltd., Tokyo, Japan) with CH3CN–10% phosphoric acid (gradient from 15:85 to 30:70) at 40 °C. The eluate at 32 min was concentrated and partitioned between ethyl acetate and H2O. The ethyl acetate layer was concentrated to dryness to yield a white powder of 4 (3.1 mg).
(2S)-2-Hydroxyheptanoyl-N-methyl-L-alanine (4)
HR-FAB-MS (m/z) found 232.1559, calcd 232.1549 [(M+H)+, C11H22NO4]; 1H NMR (600 MHz, CD3OD) δ 4.99 (1H, m, Hhaα-CH), 4.28 (1H, q, Jα,β=6.9 Hz, Alaα-CH), 2.97 (3H, s, AlaNCH3), 2.03 (1H, m, Hhaβ-CH2), 1.88 (1H, m, Hhaβ-CH2), 1.55 (3H, d, Jα,β=6.9 Hz, Alaβ-CH3), 1.47 (2H, m, Hhaγ-CH2), 1.36 (4H, m, Hhaδ-CH2 & Hhaɛ-CH2), 0.92 (3H, t, Jɛ,ζ=7.2 Hz, Hhaζ-CH3); 13C NMR (150 MHz, CD3OD) δ 170.1 (AlaC=O), 167.6 (HhaC=O), 78.5 (Hhaα-CH), 57.9 (Alaα-CH), 32.6 (Hhaδ-CH2), 32.1 (Hhaβ-CH2), 32.0 (AlaNCH3), 25.1 (Hhaγ-CH2), 23.5 (Hhaɛ-CH2), 16.5 (Alaβ-CH3), 14.3 (Hhaζ-CH3).
Ryanodine-binding assay
Cockroach membrane containing RyR was prepared as reported previously.6 The membrane of mouse leg muscle containing RyR was prepared as follows: mouse leg muscle (2.15 g) was cut and washed with 11 ml of preparation buffer (50 mM Tris-HCl, pH 8.0 containing 4 μg ml–1 each of aprotinin, leupeptin and pepstatin). It was homogenized and washed with 7 ml of the buffer. The homogenate was centrifuged (2000 g) for 10 min at 4 °C, washed and the supernatant was centrifuged again (25 000 g) for 45 min. The pellets were re-suspended in 1.7 ml of the buffer and stored at –80 °C until the binding assay.
For the binding assay, sample solutions were put into individual wells of a microplate. The samples were mixed with 10 μl ethanol solution containing 0.1 nM ryanodine, and 70 μl of the buffer, which consisted of 50 mM Tris-HCl (pH 8.0), 300 μM sucrose, 500 μM CaCl2, 1.5 M KCl, 4 μg ml–1 each of aprotinin, leupeptin and pepstatin, with 10 μl of 20 nM [9,21-3H]ryanodine (56.9 Ci mmol–1, PerkinElmer NET-950, PerkinElmer Inc, Waltham, MA, USA) solution, was also added to the microplate. The reaction was started by adding each of 10 μl of the RyR-containing membrane (2.5 mg protein per ml) described above, with the microplate subsequently being shaken for 90 min at room temperature. Free ligand was separated from the protein-bound ligand by filtration through a glass-fiber filter (PerkinElmer Printed Filtermat B) using Inotech cell harvester (Inotech Biosystems International Inc., Rockville, MD, USA), followed by washing with ice-cold 20 mM Tris-HCl buffer (pH 8.0, 3 × 300 μl). The filter was dried in a microwave oven, a solid scintillator sheet (PerkinElmer MeltiLex B/HS) was put on the filter and the sheet was melted on a hot plate. The activity retained on the filter was measured using PerkinElmer MicroBeta TriLux liquid scintillation counter. Specific binding was determined as the difference between total binding ([3H]ryanodine) and non-specific binding ([3H]ryanodine plus 500-fold excess of unlabelled ryanodine).
Other biological assays
The assay method for nematocidal and insecticidal activities was carried out using previously reported procedures.23
References
Ōmura, S. & Shiomi, K. Discovery, chemistry, and chemical biology of microbial products. Pure Appl. Chem. 79, 581–591 (2007).
Fill, M. & Copello, J. A. Ryanodine receptor calcium release channels. Physiol. Rev. 82, 893–922 (2002).
Zalk, R., Lehnart, S. E. & Marks, A.R. Modulation of the ryanodine receptor and intracellular calcium. Annu. Rev. Biochem. 76, 367–385 (2007).
Zhao, F., Li, P., Chen, S. R. W., Louis, C. F. & Fruen, B. R. Dantrolene inhibition of ryanodine receptor Ca2+ release channels. Molecular mechanism and isoform selectivity. J. Biol. Chem. 276, 13810–13816 (2001).
Xu, X., Bhat, M. B., Nishi, M., Takeshima, H. & Ma, J. Molecular cloning of cDNA encoding a Drosophila ryanodine receptor and functional studies of the carboxyl-terminal calcium release channel. Biophys. J. 78, 1270–1281 (2000).
Schmitt, M., Turberg, A., Londershausen, M. & Dorn, A. Binding sites for Ca2+-channel effectors and ryanodine in Periplaneta americana—possible targets for new insecticides. Pestic. Sci. 48, 375–388 (1996).
Ebbinghaus-Kintscher, U. et al. Phthalic acid diamides activate ryanodine-sensitive Ca2+ release channels in insects. Cell Calcium 39, 21–33 (2006).
Cordova, D. et al. Anthranilic diamides: a new class of insecticides with a novel mode of action, ryanodine receptor activation. Pestic. Biochem. Physiol. 84, 196–214 (2006).
Ōmura, S., Shiomi, K. & Masuma, R. Substance FKI-1033 and process for producing the same (Kitasato Institute). U S Patent 7,541,336 (2009).
Monma, S. et al. Verticilide: elucidation of absolute configuration and total synthesis. Org. Lett. 8, 5601–5604 (2006).
Domsch, K. H., Gams, W. & Anderson, T.- H. Compendium of Soil Fungi 2nd edn (IHW, Verlag, Eching, 2007).
Kanaoka, M . et al. Bassianolide, a new insecticidal cyclodepsipeptide from Beauveria bassiana and Verticillium lecanii. Agric. Biol. Chem. 42, 629–635 (1978).
Ohyama, M. Chemistry of anthelmintic cyclodepsipeptides. Sci. Rep. Meiji Seika Kaisha 45, 8–34 (2006).
Taber, W. A. & Vining, L. C Amidomycin, a new antibiotic from a Streptomyces. Production, isolation, assay, and biological properties. Can. J. Microbiol. 3, 953–965 (1957).
Pettit, G. R. et al. Antineoplastic agents. Part 409: isolation and structure of montanastatin from a terrestrial actinomycete. Bioorg. Med. Chem. 7, 895–899 (1999).
Suzuki, A. et al. Bassianolide, a new insecticidal cyclodepsipeptide from Beauveria bassiana and Verticillium lecanii. Tetrahedron Lett. 2167–2170 (1977).
Sasaki, T. et al. A new anthelmintic cyclodepsipeptide, PF1022A. J. Antibiot. 45, 692–697 (1992).
Sasaki, T. et al. Cyclic depsipeptide and production thereof (Meiji Seika Kaisha, Ltd.). Japanese Patent Application JP 1993-170749 (1993).
Ohyama, M. et al. Cyclic depsipeptide and its production (Meiji Seika Kaisha, Ltd.). Japanese Patent Application JP 1994-184126 (1994).
Ohyama, M. et al. Process for producing cyclodepsipeptide compounds and novel cyclodepsipeptide (Meiji Seika Kaisha, Ltd.). PCT Int. Appl. WO 98/05655 (1998).
Nakajyo, S. et al. On the inhibitory mechanism of bassianolide, a cyclodepsipeptide, in acetylcholine-induced contraction in guinea pig taenia coli. Jpn. J. Pharmacol. 33, 573–582 (1983).
Welz, C., Harder, A., Schnieder, T., Hoglund, J. & von Samson-Himmelstjerna, G. Putative G protein-coupled receptors in parasitic nematodes — potential targets for the new anthelmintic class cyclooctadepsipeptides? Parasitol. Res. 97, S22–32 (2005).
Enomoto, Y. et al. Isolation of a new antibiotic oligomycin G produced by Streptomyces sp. WK-6150. J. Antibiot. 54, 308–313 (2001).
Acknowledgements
We are grateful to Ms Akiko Nakagawa and Ms Chikako Sakabe of the School of Pharmacy, Kitasato University, for measurements of mass spectra. This work was supported by a Grant-in-Aid for Scientific Research (17590091). This work was also supported in part by the Grant of the 21st Century COE Program, Ministry of Education, Culture, Sports, Science, and Technology (MEXT).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Shiomi, K., Matsui, R., Kakei, A. et al. Verticilide, a new ryanodine-binding inhibitor, produced by Verticillium sp. FKI-1033. J Antibiot 63, 77–82 (2010). https://doi.org/10.1038/ja.2009.126
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2009.126
Keywords
This article is cited by
-
Decoding and reprogramming fungal iterative nonribosomal peptide synthetases
Nature Communications (2017)
-
New verticilides, inhibitors of acyl-CoA:cholesterol acyltransferase, produced by Verticillium sp. FKI-2679
The Journal of Antibiotics (2012)