Abstract
The oncoprotein c-Myc is a key transcription factor with essential functions in the nucleolus (NO) to regulate ribosomal RNA (rRNA) synthesis, ribosome biogenesis, and cell proliferation. Yet, the mechanism that regulates the distribution and function of nucleolar c-Myc is still not completely understood. In this study, we identified nucleolar protein ENBA1 binding protein 2 (EBP2) as a novel functional binding partner of c-Myc. We found that coexpression of EBP2 markedly relocalized c-Myc from the nucleus to the NO, whereas depletion of EBP2 reduced the nucleolar distribution of c-Myc. Further study indicated that EBP2 is a direct binding partner of c-Myc and can block the degradation of c-Myc in a FBW7 (F-box and WD repeat domain containing 7)-independent manner. Moreover, EBP2 is a transcriptional target of c-Myc. c-Myc can bind to the promoter of EBP2 and positively regulate the EBP2 expression. Both protein and mRNA levels of EBP2 are upregulated in lung cancer samples and positively correlated with c-Myc expression. Functionally, EBP2 promotes c-Myc-mediated rRNA synthesis and cell proliferation. Collectively, our study indicates that EBP2 is a novel binding partner of c-Myc that regulates the function of nucleolar c-Myc, cell proliferation, and tumorigenesis via a positive feedback loop.
Similar content being viewed by others
Main
c-Myc is a key transcriptional factor that has critical functions in various biological processes such as cell growth, metabolism, angiogenesis, and DNA repair.1, 2 Moreover, c-Myc is a potent oncoprotein that is frequently deregulated in many human cancers and can promote tumorigenesis in a very wide variety of tissues.1, 3 Recent studies indicate that c-Myc is a universal transcriptional amplifier of expressed genes.4, 5 c-Myc is mainly localized in the nucleus in cells to regulate the transactivation or repression of various genes by direct DNA binding.6 Except nuclear distribution, c-Myc is found in the cytosol and the nucleolus (NO) in cells.2, 7 Myc-Nick, a truncated form of Myc, is localized in the cytosol and regulates cell differentiation.7 Nucleolar-localized c-Myc is essential for ribosome biogenesis by binding to ribosomal DNA (rDNA), activating RNA polymerase I, and regulating the transcription of rDNA.8, 9 The function of c-Myc in the NO is tightly controlled by protein degradation and its binding proteins.10 Yet, SCF-Fbw7γ is the major reported E3 ubiquitin ligase to degrade c-Myc in the NO via ubiquitin–proteasome system.11, 12 Recent study indicates that nucleophosmin (NPM) is required for the nucleolar localization of c-Myc and promotes the degradation of c-Myc in the nucleoli in a Fbw7-independent manner.10 However, the underlying mechanisms that regulate c-Myc localization and functions in the nucleoli remain to be investigated.
ENBA1 binding protein 2 (EBP2) is originally identified as a binding protein of Epstein–Barr virus (EBV) nuclear antigen 1 that is important for EBV segregation.13, 14 Yeast homolog EBP2 (Ebp2p) is an essential nucleolar protein required for pre-ribosomal RNA (rRNA) processing and ribosomal subunit assembly.15, 16, 17, 18 In mammalian cells, EBP2 interacts with nucleostemin and is localized to the NO.19 Moreover, human EBP2 is associated with chromosome in mitosis.20 Overexpression of EBP2 has been shown to be able to promote the cell proliferation and chromosome instability of 293 cells or NIH3T3 cells.21, 22 A recent study demonstrates that EBP2 is essential for the nucleolar localization of Fbw7γ via direction interaction.23 However, the biological functions of mammal EBP2, such as whether it may affect the stability of Fbw7γ substrates, remain not completely understood.
In this study, we identified EBP2 as a novel binding partner of c-Myc. We found that EBP2 can relocalize c-Myc to the NO, block the c-Myc degradation, and promote the expression of rRNA. Moreover, EBP2 is a direct transcriptional target of c-Myc. Thus, EBP2 and c-Myc forms a positive feedback regulation loop that promotes cancer cell proliferation. We also found that EBP2 is upregulated in lung cancer tissues where its expression is highly correlated with c-Myc. Thus, our study uncovered a novel function of EBP2 to regulate cancer cell proliferation through c-Myc and identified that EBP2 may be a novel therapeutic target to cancers.
Results
Relocalization of c-Myc into the NO by EBP2
c-Myc is a key transcriptional factor with important functions in the nucleoli.2, 8, 9 Its stability and activity in the nucleoli are tightly controlled by its binding partners such as E3 ubiquitin ligase Fbw7γ and NPM.10, 11 Recent study showed that EBP2 is a Fbw7γ pseudosubstrate and is essential for nucleolar localization of Fbw7γ.23 However, it is still unclear whether EBP2 affects the stability/function of c-Myc. To this end, we coexpressed HA-c-Myc with EBP2 in HeLa cells. Our data from an immunofluorescence assay showed that c-Myc is hardly detected in the NO when expressed alone (Figure 1a). Consistent with previous report,10 treatment with proteasome inhibitor MG132 resulted in the nucleolar localization of c-Myc in 17% cells (Figure 1a and Supplementary Figure S1A). Surprisingly, we found that when coexpressed with EBP2, c-Myc was translocated into the NO where it is well colocalized with EBP2 in 30% cells (Figure 1a). Moreover, treatment with MG132 further increased the nucleolar distribution of c-Myc. This result suggests that EBP2 may redistribute c-Myc from the nucleoplasm (NP) to the NO independent of protein degradation. To confirm this result, we used a density-gradient centrifugation-based cell fraction assay to separate the whole-cell extracts into the NO, NP, and cytoplasm. As shown in Figure 1b, the amount of c-Myc in the NO was significantly increased in the presence of EBP2. Conversely, knockdown of EBP2 markedly reduced the amount of endogenous c-Myc in the NO (Figure 1c). Collectively, these data suggest that EBP2 is a positive regulator of the nucleolar distribution of c-Myc.
EBP2 redistributed c-Myc into the nucleolus. (a) c-Myc was redistributed into the nucleolus when EBP2 was coexpressed. HA-c-Myc was transfected into HeLa cells with or without FLAG-EBP2. The cells were treated with or without MG132, and subcellular localization of c-Myc and EBP2 was analyzed using immunofluorescence assay. The ratio of cell with nucleolus localized c-Myc was measured (around 200 cells were counted for each panel). (b) Cell fractions of HEK293T cells transfected with empty vector or EBP2 were analyzed using western blotting. WCL, whole-cell lysate; NO, nucleolus; NP, nucleoplasm; CP, cytoplasm. Dilution: WCL:NO:NP:CP=1:5:1:1. The relative levels of c-Myc were quantified using Image J. (c) Cell fractions of A549 cells depleted with EBP2 were analyzed using western blotting. Dilution: WCL:NO:NP:CP=1:5:1:1. The relative levels of endogenous c-Myc were quantified using Image J. *P<0.05 was deemed statistically significant
c-Myc binds to EBP2 directly
Several studies indicated that the subcellular localization of c-Myc can be changed by its binding partner. Thus, we examined whether EBP2 is a binding partner of c-Myc. Our data from co-immunoprecipitation (IP) assay showed that c-Myc and EBP2 interacted with each other in the transfected cells (Figure 2a and Supplementary Figure S2A). Endogenous EBP2 was also able to be co-purified with endogenous c-Myc in A549 cell (Figure 2b). Previous study indicates that EBP2 binds to Fbw7 through a CPD motif in EBP2 N-terminus. To test the possibility that EBP2 binds c-Myc through Fbw7, we mutated the CPD of EBP2 (EBP2-T3A) and its interaction with c-Myc was examined. Our data showed that both wild-type and T3A mutant EBP2 efficiently bound to c-Myc (Figure 2c), suggesting that binding EBP2 to c-Myc is independent of Fbw7. Their direct interaction was also examined using a GST pull-down assay. Our data showed that purified c-Myc can associate with purified GST-EBP2 as well (Figure 2d). Taken together, these results indicate that EBP2 is a direct binding partner of c-Myc.
Binding of EBP2 to c-Myc. (a) c-Myc was transfected into HEK293T cells with or without FLAG-EBP2. Immunoprecipitation assay was applied 36 h after transfection using an anti-FLAG antibody. The interaction was analyzed using western blotting. (b) Endogenous c-Myc was immunoprecipitated using an anti-c-Myc antibody, irrelevant rabbit IgG was used as control. The interaction was analyzed using western blotting. (c) Empty vector, wild-type (WT) or T3A-mutant (T3A) FLAG-EBP2 was transfected with c-Myc into HEK293T cells. Immunoprecipitation assay was applied using an anti-FLAG antibody. The interaction was analyzed using western blotting. (d) Purified GST or GST-EBP2 proteins were incubated with cell lysates expressing c-Myc. The interaction was analyzed using western blotting. (e) The domains of EBP2 required for its interaction with c-Myc was analyzed using an immunoprecipitation assay and western blotting. (f) The domains of c-Myc required for its interaction with EBP2. A, 1-360; B, 1-150; C, 143-439; and D, 143-360. The interaction was analyzed using an immunoprecipitation assay and western blotting
Next, we examined the molecular basis of their interaction. Our data from co-IP assay showed that the N-terminal but not C-terminal domain of EBP2 bound to c-Myc (Figure 2e). Interestingly, both N-terminal and C-terminal domains of c-Myc could bind to EBP2 (Figure 2f). This result was confirmed by the immunofluorescence assay showing that EBP2 could recruit the deletion mutants of c-Myc containing either N-terminus or C-terminus to the nucleoli (Supplementary Figure S2B). These data indicate that N-terminus of EBP2 binds to the multiple sites of c-Myc.
EBP2 stabilizes c-Myc independent of Fbw7
It has been reported that c-Myc is degraded in the nucleoli.10 As EBP2 can relocalize c-Myc to the nucleoli, we thus measured whether EBP2 affects c-Myc stability. To this end, we co-expressed c-Myc with an increasing amount of EBP2 in HEK293T cells and the protein levels of c-Myc were detected. As shown in Figure 3a, the protein levels of c-Myc were increased on EBP2 coexpression. Moreover, coexpression with EBP2 prolonged the half-life of c-Myc when measured using a CHX-chase assay (Figure 3b). Conversely, the protein levels of c-Myc, but not β-catenin or IκB-α, were decreased on EBP2 knockdown by two specific small interfering RNA (siRNA) in both A549 and H1299 cells (Figure 3c), indicating that c-Myc protein was specifically regulated by EBP2 via degradation. In contrast, the mRNA levels of c-Myc were not significantly affected by EBP2 knockdown (Supplementary Figure S3A). Knockdown of EBP2 also promoted the turnover of endogenous c-Myc proteins as measured using a CHX-chase assay (Figure 3d). Treatment with proteasome inhibitor MG132 for 6 h completely abolished the effect of EBP2 knockdown on c-Myc protein levels (Figure 3e). Collectively, these results indicate that EBP2 can block the degradation of c-Myc via a proteasome activity-dependent manner rather than protein synthesis.
EBP2 stabilized c-Myc. (a) HA-c-Myc was expressed in HEK293T cells with vector or an increasing dose of FLAG-EBP2. The protein levels of c-Myc were analyzed using western blotting. (b) HA-c-Myc was co-expressed with or without EBP2 in HEK293T cells. Cells were then treated with CHX for the indicated time. The expression of c-Myc, EBP2, and actin were determined using western blotting. (c) A549 cells or H1299 cells were transfected with siNC or two siRNAs against EBP2: siEBP2-1 (siE1), siEBP2-2 (siE2) for 72 h. The protein levels of c-Myc, EBP2, β-catenin, IκB-α and actin were determined using western blotting. The relative levels of c-Myc and EBP2 were quantified using Image J. (d) A549 cells were transfected with siNC, siEBP2-1, and siEBP2-2 for 72 h. Cells were then treated with CHX for indicated times. The expression of c-Myc, EBP2, and actin were determined using western blotting. The relative level of c-Myc was quantified by Image J. (e) A549 cells were transfected with siNC, siEBP2-1 and siEBP2-2 for 72 h. Cells were divided into two wells and one well was treated with proteasome inhibitor MG132 (10 μM) or DMSO as control for 8 h before collection
EBP2 has been shown to be able to recruit Fbw7γ to the nucleoli, which is a well-established E3 ubiquitin ligase for c-Myc.11, 30, 31 Thus, there is a possibility that EBP2 stabilizes c-Myc through blocking the binding of Fbw7 to c-Myc. To test whether the effect of EBP2 on c-Myc is dependent on FBW7 (F-box and WD repeat domain containing 7), EBP2 was depleted in A549 cells together with or without Fbw7 knockdown using a specific siRNA against all three isoforms of Fbw7, and the protein levels of c-Myc were examined. As expected, knockdown of Fbw7 significantly increased the protein levels of c-Myc (Supplementary Figure S3A). However, knockdown of EBP2 still reduced the c-Myc protein levels in Fbw7-depleted cells (Supplementary Figure S3B). The CHX-chase assay indicated that EBP2 regulated the c-Myc turnover in a Fbw7-independent manner (Supplementary Figure S3C). Moreover, both wild-type and CPD-mutated form of EBP2 (EBP2-3TA) could stabilize c-Myc (Supplementary Figure S3D). These results together indicate that EBP2 affects the c-Myc stability independent of Fbw7.
c-Myc regulates EBP2 mRNA expression
During our experiment, we also noticed that knockdown of c-Myc markedly reduced the protein levels of EBP2 (Figure 4a). As a transcription factor, c-Myc is known to regulate the expression of numerous genes.1, 4 Thus, we asked whether c-Myc affects the mRNA level of EBP2. As shown in Figure 4b, knockdown of c-Myc reduced the mRNA levels of EBP2 and overexpression of c-Myc significantly increased the mRNA levels of EBP2 (Figure 4c). These results indicate that c-Myc is a positive regulator of EBP2 mRNA transcription.
c-Myc regulated EBP2 expression at transcription level. (a) A549 cells were transfected with siRNAs as indicated for 72 h. The expression of EBP2, c-Myc, and actin was analyzed using western blotting. (b) A549 cells were transfected with as indicated. The mRNA level of EBP2 was measured using qPCR. (c) c-Myc was overexpressed in HEK293T cells and the mRNA level of EBP2 was measured using qPCR. (d) The 2-kb promoter of EBP2 contains four predicted c-Myc binding site. (e) Binding of c-Myc to the EBP2 promoter. H1299cells were subjected to ChIP assay using an anti-c-Myc antibody. The binding of c-Myc to the promoter was measured using qPCR. Five pairs of primers were used. (f) One kilobase promoter of EBP2 was cloned into pGL3 vector. Empty vector and an increasing dose of c-Myc was transfected into HEK293T cells with Renilla. The luciferase activity of the report gene was measured. (g) c-Myc promotes the EBP2 promoter activity of EBP2. Wild-type or mutant EBP2 promoter luciferase reporter gene was cotransfected with or without c-Myc in the HEK293T cells and luciferase activity was measured. *P<0.05 was deemed statistically significant and **P<0.01 highly significant
As a transcription factor, c-Myc can regulate the gene expression via direct binding to the promoter of targets via an E-box sequence.32 To further examine whether c-Myc can affect the EBP2 mRNA at transcriptional level, we analyzed the EBP2 promoter using Transcription Element Search System (http://www.cbil.upenn.edu/tess) and identified four conserved c-Myc binding sequences in EBP2 promoter (Figure 4d). Therefore, we examined whether c-Myc can directly bind to these E-box sequence of EBP2 using a ChIP assay. We designed five pairs of primers in the 2-kb promoter region corresponding to the four predicted binding sites, and one locates in the region without E-box sequence as control (Figure 4f). Our data showed that c-Myc can strongly bind to the third E-box and with a weaker affinity to the fourth E-box, but not other regions (Figure 4c and data not shown).
To further confirm that c-Myc is a transcription factor of EBP2, we employed a luciferase-based reporter gene system. The 1-kb promoter from start codon of EBP2 was cloned into pGL3 vector and coexpressed with empty vector or with an increasing amount of c-Myc in HEK293T cells. Our data showed that the report gene of EBP2 promoter was activated by c-Myc in a dosage-dependent manner (Figure 4f). Simultaneous mutation of the third and fourth E-box significantly abolished the c-Myc-mediated transactivation of EBP2 promoter (Figure 4g). Taken together, we conclude that c-Myc can promote EBP2 gene expression by direct binding to its promoter via conserved E-box DNA-binding sites.
EBP2 overexpression promotes cell proliferation and tumorigenesis in vivo
c-Myc is a critical transcriptional factor that can regulate cell proliferation.5 Thus, we examined whether EBP2 may also regulate cancer cell proliferation. We first measured the correlated expression of EBP2 with c-Myc in various lung cancer cells. As show in Figure 5a, the protein levels of both EBP2 and c-Myc were relatively low in normal human lung epithelial cell line BEAS-2B. In contrast, EBP2 was highly expressed in most lung cancer cells that we examined (Figure 5a). Importantly, the protein levels of c-Myc were positively correlated with the levels of EBP2 in cancer cell lines (Figure 5a).
EBP2 promoted c-Myc-dependent cell proliferation and tumorigenesis. (a) Protein levels of c-Myc and EBP2 were detected in the normal lung cells and lung cancer cell lines using western blotting. (b) Overexpressed EBP2 promoted cell proliferation similar to c-Myc. Cell proliferation of H1299 cells were measured using a MTT assay. (c) Knockdown of EBP2 inhibited cell proliferation. Cell proliferation ofA549 cells were measured using a MTT assay. (d) Knockdown of EBP2 inhibited cell proliferation of A549 and H1299 cells measured using a colony formation assay. (e) EBP2 promoted cell proliferation via a c-Myc-dependent manner. H1299 cells were transfected as indicated and cell proliferation was measured using a MTT assay. (f) Overexpressed EBP2 promoted H1299 tumorigenesis. Stable cell lines of H1299 overexpressing empty vector or EBP2 was injected into nude mouse (5 × 106 cells per mice). The diameter of the tumor was measured every 2 days from 16th day after injection. The tumors were obtained after 28 days after injection as shown
We next examined whether EBP2 can affect cancer cell proliferation. EBP2 and c-Myc were overexpressed in H1299 cells, and cell proliferation was measured using an MTT assay. As shown in Figure 5b, overexpression of EBP2 can significantly promote H1299 cell growth similar to c-Myc. EBP2 was also depleted using two different siRNAs in A549 cells and the cell proliferation was measured. Our data showed that cell proliferation was sharply repressed on EBP2 knockdown (Figure 5c). This result was confirmed using a colony formation assay (Figure 5d). To examine whether the promotion of cell proliferation by EBP2 is dependent on c-Myc, we overexpressed EBP2 with or without c-Myc depletion in H1299 cells. Our results showed that EBP2 can hardly promote H1299 proliferation when c-Myc was deleted (Figure 4e).
We then examined whether EBP2 promoted tumorigenesis. EBP2 was stably expressed in H1299 cells, and cells stably expressing EBP2 or empty vector were injected subcutaneously into nude mouse. The tumorigenesis was measured. Our result showed that overexpression of EBP2 strongly promoted tumorigenesis (Figure 5f). Taken together, our data indicate that EBP2 is required for cell growth of lung cancer cells and can promote tumorigenesis in nude mice.
The levels of EBP2 are positively correlated with c-Myc in human lung cancer tissues
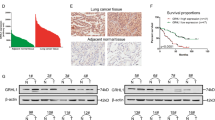
c-Myc is an oncoprotein and is highly expressed in various cancers cells, including lung cancers.33, 34 Our data indicated that EBP2 can promote tumorigenesis. Thus, we measured the expression levels of EBP2 in lung cancer tissues using an IHC assay. The specificity of EBP2 antibody was confirmed by IF and IHC analysis (Supplementary Figure S4). EBP2 was stained in both carcinoma adjacent tissues and lung cancer tissues. Our data showed that the expression levels of EBP2 were significantly higher in lung cancer tissues than in adjacent tissues (Figure 6a). Moreover, through staining of serial sections from the same tumor samples, we found that the expression levels of EBP2 were positively correlated with c-Myc in lung cancer tissue (Figures 6b and c). This is consistent with our conclusion that EBP2 is a transcriptional target of c-Myc. These data indicated that the expression levels of EBP2 are correlated with c-Myc in lung cancer.
Protein levels of EBP2 were upregulated and positively correlated with c-Myc in human lung cancer tissues. (a) Expression of EBP2 in carcinoma adjacent tissues and lung cancer tissues were examined using an immunohistochemical staining assay of two lung cancer patient samples. (b) The protein levels of EBP2 were positively correlated with c-Myc. Serial sections of lung cancer samples were stained with EBP2 and c-Myc separately using an immunohistochemical staining assay. Representative examples were shown. (c) Statistical analysis of the correlation between the protein levels of EBP2 and c-Myc in the lung cancer tissue array
EBP2 enhances c-Myc-mediated rDNA transcription
It has been reported that c-Myc can stimulate the rDNA transcription, which is essential for cell proliferation.8, 9 Thus, we examined whether EBP2 can affect rRNA synthesis mediated by c-Myc. To this end, we overexpressed c-Myc or EBP2 in HEK293T cells, and the rRNA levels were measured using qRT-PCR. Our data showed that the rRNA levels were markedly increased in both c-Myc- and EBP2-transfected cells (Figure 7a). Conversely, knockdown of either c-Myc or EBP2 in A549 cells dramatically reduced the rRNA levels (Figure 7b), suggesting that EBP2 is a regulator of rRNA synthesis. Moreover, coexpression of EBP2 significantly increased the c-Myc-promoted rRNA levels in HeLa cells (Figure 7c). This indicates that EBP2 is a positive regulator of c-Myc-mediated rRNA synthesis.
EBP2 promoted rDNA transcription. (a) HEK293T cells were transfected with empty vector, c-Myc, or EBP2 for 24 h. The levels of rRNA were examined using qPCR. (b) EBP2 and c-Myc were knocked down in A549 cells and the levels of rRNA were examined using qPCR. (c) HeLa cells infected with lentivirus-encoding vector or Flag-c-Myc were transfected with empty vector or EBP2. The cells were starved for 48 h The levels of rRNA were examined using qPCR
Discussion
c-Myc has essential functions in the NO to regulate the synthesis of rRNA, RNA polymerase I activity, and cell proliferation.35 In this study, we have identified EBP2 as a novel nucleolar regulator of c-Myc. We found that EBP2 can directly bind to c-Myc and regulate the c-Myc nucleolar localization, cell proliferation, and rRNA transcription. Moreover, we found that c-Myc positively regulates the expression of EBP2 via directly binding to the EBP2 promoter. Thus, our study suggests a positive feedback loop to regulate the c-Myc function by EBP2.
Although it is well known that c-Myc is able to be localized in the nucleoli, the mechanism that controls the c-Myc nucleolar localization is still not completely understood. In our study, we found that overexpression of EBP2 can also relocalize c-Myc to the NO. Importantly, depletion of EBP2 reduced the nucleolar distribution of endogenous c-Myc, suggesting that EBP2 is also required to keep the protein levels of c-Myc in the nucleoli. Our study indicated that EBP2 can directly bind to c-Myc, providing possible mechanism that EBP2 recruit c-Myc to the nucleoli via direction interaction. Recent study provided clear evidence to show that nucleolar protein NPM is essential for c-Myc nucleolar localization.10 However, it has been shown that EBP2 cannot interact with NPM and the distribution of EBP2 is not affected by NPM,23 suggesting that recruitment of c-Myc to the nucleoli by EBP2 is not possibly through the NPM. Thus, binding to EBP2 may be a novel mechanism that recruits c-Myc to the nucleoli.
It is known that c-Myc is degraded rapidly in the nucleoli.11 Yet, E3 ubiquitin ligase Fbw7γ is the only reported E3 to promote the c-Myc degradation in the nucleoli.11 Whether there is any other mechanism to regulate the c-Myc stability is not clear. However, recent study indicates that an unknown mechanism may exist to control c-Myc degradation in the nucleoli.10 In our current study, we found that EBP2 can stabilize c-Myc through the Fbw7-independent way. Although the exact mechanism that controls c-Myc degradation is still unclear, our study suggests that EBP2, together with other nucleolar regulator of c-Myc, such as NPM, may cooperatively keep the protein levels of c-Myc in the NO. Deregulated expression of nucleolar binding partner of c-Myc may promote tumorigenesis.
Although previous study indicates that overexpression of EBP2 can promote the cell proliferation of HEK293 cells,21 its role in the tumorigenesis is still unclear. In our study, we identified that EBP2 is upregulated in lung cancer cells and tissues. Moreover, EBP2 is required for cell proliferation. Importantly, we found that EBP2 is a c-Myc binding partner of c-Myc and required for keeping the amount of c-Myc in nucleoli. Thus, our study uncovers a novel function of EBP2 in cancers and identified EBP2 as a potential oncoprotein.
In summary, we identified EBP2 as a novel nucleolar regulator of c-Myc and demonstrated that EBP2 may regulate c-Myc nucleolar localization, stability, and cell proliferation. On the other hand, c-Myc positively regulates EBP2 expression at the transcriptional level. Thus, EBP2 and c-Myc forms a positive feedback regulatory loop (Figure 7d), which has important roles in cell proliferation and tumorigenesis.
Materials and Methods
Plasmids and cell culture
The expression plasmid of EBP2 was cloned into pcDNA3.1 vector with a FLAG tag at the C-terminus and c-Myc was cloned into pcDNA3.1 vector with an HA tag at the N-terminus by standard cloning methods. All vectors were confirmed by DNA sequencing. HEK293T and HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco, Mount Waverley, VIC, Australia) supplemented with 10% heat-inactivated fetal bovine serum (FBS) at 37 °C in 5% CO2. Human lung epithelial cell BEAS-2B and human lung cancer cells, including H358, H441, PC9, H197, H1299, A549, H460, SPC-A1, and 95D cells, were cultured in 1640 medium (Gibco) supplemented with 10% heat-inactivated FBS. Transfections were performed using calcium phosphate–DNA coprecipitation for 293T cells and SunbioTrans-EZ for HeLa cells (Shanghai Sunbio Medical Biotechnology Co., Ltd, Shanghai, China).
Small interfering RNA
A549 and H1299 cells were transfected with siRNA oligonucleotides using Lipofectamine. Two different oligonucleotides siEBP2-1 (273: 5′-GGACCAGAAAGCTGTT GATCCAGAA-3′) and siEBP2-2 (640: 5′-GAGGGAGATCAGAAACCTCTGGCAC-3′) against EBP2 were used. Oligonucleotides against FBW7 were used as described previously.24
Antibodies, immunoprecipitation, and western blotting
IP and western blotting were performed as our previous report.24 Briefly, cells were transfected, treated with 10 μM MG132 for 6 h, and lysed using 2 × RIPA buffer (Tris-HCl, pH 7.4 (100 mM), NaCl (300 mM), 1% NP-40, 2% sodium deoxycholate, 10 mM NaF, and 10 mM Na vanadate). The cell lysates were cleared by centrifugation and incubated with 1 μg antibody for 1 h at 4 °C followed by incubation with 15 μl protein A and G beads (Santa Cruz, Santa Cruz, CA, USA) for 2 h at 4 °C. Immunoprecipitates were subjected to western blotting. For western blotting analysis, cells were scraped from the dishes into the lysis buffer. A total of 25 mg of total protein was separated by SDS-PAGE and blotted with anti-c-Myc (Epitomics, Burlingame, CA, USA), anti-NPM (Epitomics), anti-EBP2 (Abnova, Taipei City, Taiwan, China), anti-β-catenin (CST, Danvers, MA, USA), anti-IκB-α (CST), anti-α-tubulin (Abmart, Shanghai, China), anti-actin (Abmart), or anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Abmart).
Ethics statement
Human lung cancer specimens and carcinoma adjacent area specimens were collected from the Fudan University Shanghai Cancer Center. All specimens were encoded to protect patients under protocols approved by the Institutional Review Board of the Fudan University Cancer Center. All participants gave written informed consent.25
Immunofluorescence
Immunofluorescence was performed as described previously.24 Briefly, cells cultured on coverslips were transfected and fixed with 4% paraformaldehyde, permeabilized using 0.1% Triton X-100, blocked with 1% BSA in PBS for 1 h, and stained with corresponding antibodies. The slides were then stained with Texas Red-labeled anti-rabbit secondary antibody and FITC-labeled anti-mouse secondary antibody (1:100; Invitrogen, Carlsbad, CA, USA). The nuclei were stained with 0.5 μg/ml 4′,6-diamidino-2-phenylindole (Sigma, St. Louis, MO, USA). Images were analyzed using a Leica SP5 confocal microscope (East China Normal University, Shanghai, China).
Nucleolar isolation
This experiment was performed according to the Lamond’s protocol.26, 27 Briefly, ∼106 cells were collected, washed twice with PBS, and resuspended in 1 ml buffer (10 mM HEPES–KOH, pH 7.9, 1.5 M MgCl2, 10 mM KCl, and 0.5 mM DTT) for 30 min on ice. Phenylmethylsulfonyl fluoride was added to a final concentration of 0.2 mM, and the mixture was then Dounce homogenized until all cytoplasmic membranes were disrupted. For cytosolic isolation, cells were centrifuged at 228 × g for 5 min at 4 °C to obtain the supernatant. The resulting pellet was resuspended in 0.45 ml of 0.25 M sucrose–10 mM MgCl2, layered onto 0.45 ml of 0.35 M sucrose–0.5 mM MgCl2, and centrifuged at 1430 × g for 5 min at 4 °C. Pelleted nuclei were then resuspended in 0.75 ml of 0.35 M sucrose–0.5 mM MgCl2 with protease and phosphatase inhibitors. Nuclei were sonicated to disrupt the nuclear membrane. The nuclear isolate was layered on the top of 0.75 ml of 0.88 M sucrose–0.5 mM MgCl2 and centrifuged at 2800 × g for 10 min at 4 °C. The pellet was resuspended in 0.5 ml of 0.35 M sucrose–0.5 mM MgCl2, and sucrose layering was repeated as described above. Nucleoli were fractionated as the subsequent pellet.
RNA extraction and qPCR
Total mRNA was isolated using TRIzol (Invitrogen) and 500 ng RNA were used to synthesize cDNA using PrimeScriptTM RT reagent kit (Takara, Dalian, China, DRR037A) according to the manufacturer’s protocol. qPCR was performed in duplicates using the SYBR Premix Ex Taq (Takara, DRR420A) on an M x 3000P System (Stratagene, La Jolla, CA, USA). Data were calculated according to the ΔCt relative quantification method. The primers are as follows: c-Myc sense primer 5′-TCAAGAGGCGAACACACAAC-3′, antisense primer 5′-GGCCTTTTCATTGTTTTCCA-3′; EBP2 sense primer 5′-AGAAAGGCTCAAAGTGGAACA-3′, antisense primer 5′-CATGACACCAACAGAAGGGAT-3′; rDNA sense primer 5′-GCCTTCTCTAGCGATCTGAGAG-3′, antisense primer 5′-CCATAACGGAGGCAGAGACA-3′; and GAPDH sense primer 5′-TGACAACTTTGGTATCGTGGAAG-3′, antisense primer 5′-CAGTAGAGGCAGGGATGATGTT-3′.
Chromatin immunoprecipitation PCR
Chromatin immunoprecipitation (ChIP) analysis was performed as described.9, 28 In brief, ∼107 cells were resuspended in 10 ml warm medium containing 1% formaldehyde for 10 min at room temperature. Cells were lysed in lysis buffer (1% SDS, 50 mM Tris (pH 8.0), 10 mM EDTA, 10 mM Na-butyrate, and protease inhibitor cocktail) and sonicated 15 times for 15 s to achieve a chromatin size of 100–500 bp. c-Myc was immunoprecipitated using c-Myc antibody (Epitomics). The extracted DNA from each IPwas resuspended in 50 μl of TE (10 mM Tris (pH 8.0), 1 mM EDTA). In parallel, an input DNA sample was prepared and resuspended in 100 μl TE. Real-time PCR was used to quantitate the immunoprecipitated DNA relative to a standard curve built with 1 μl of input dilutions. The fold enrichment was calculated relative to the background detected with nonspecific rabbit IgG for each primer set.
MTT assay
A549 or H1299 cells transfected with plasmids or siRNA for 48 h were seeded in 96-well plates at an initial cell density of 500 cells per well, MTT (3-[4,5-dimethylthiozol-2-yl]-2,5diphenyltetrazoliumbromide) (T0793, Sangon Biotech, Shanghai, Co., Ltd, Shanghai, China) was added to each well for 4 h. The reaction was stopped by removing medium with MTT and adding 150 μl DMSO to each well. The absorbance was measured at 490 nm using a microplate spectrophotometer (SpectRA MAX190, Molecular Devices Corp., Sunnyvale, CA, USA).
Colony formation assay
A549 or H1299 cells were transfected siRNA and seeded in 6-well plates at an initial cell density of 200 cells per well. Colonies were allowed to form for 2 weeks and stained with 0.05% crystal violet after methanol fixation.29
Tumor xenografts
Six-week-old male nude mice were obtained from Shanghai Experimental Animal Center (Shanghai, China). H1299-EBP2 (5 × 106) stable cells and control cells were subcutaneously injected, respectively, into the right side and left dorsal flanks of each nude mouse. From 16 day after injection, the diameter of the tumor was measured every 2 days by a vernier caliper. Twenty-eight days after injection, the mice were killed and the tumors were photographed.
Abbreviations
- EBP2:
-
ENBA1 binding protein 2
- FBW7:
-
F-box and WD repeat domain containing 7
- NPM:
-
nucleophosmin
- GAPDH:
-
glyceraldehyde-3-phosphate dehydrogenase
- rDNA:
-
ribosomal DNA
- rRNA:
-
ribosomal RNA
References
Meyer N, Penn LZ . Reflecting on 25 years with MYC. Nat Rev Cancer 2008; 8: 976–990.
van Riggelen J, Yetil A, Felsher DW . MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 2010; 10: 301–309.
Pelengaris S, Khan M, Evan G . c-MYC: more than just a matter of life and death. Nat Rev Cancer 2002; 2: 764–776.
Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 2012; 151: 56–67.
Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 2012; 151: 68–79.
Knoepfler PS . Myc goes global: new tricks for an old oncogene. Cancer Res 2007; 67: 5061–5063.
Conacci-Sorrell M, Ngouenet C, Eisenman RN . Myc-nick: a cytoplasmic cleavage product of Myc that promotes alpha-tubulin acetylation and cell differentiation. Cell 2010; 142: 480–493.
Arabi A, Wu S, Ridderstrale K, Bierhoff H, Shiue C, Fatyol K et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol 2005; 7: 303–310.
Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN et al. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol 2005; 7: 311–318.
Li Z, Hann SR . Nucleophosmin is essential for c-Myc nucleolar localization and c-Myc-mediated rDNA transcription. Oncogene 2013; 32: 1988–1994.
Welcker M, Orian A, Grim JE, Eisenman RN, Clurman BE . A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr Biol 2004; 14: 1852–1857.
Bonetti P, Davoli T, Sironi C, Amati B, Pelicci PG, Colombo E . Nucleophosmin and its AML-associated mutant regulate c-Myc turnover through Fbw7 gamma. J Cell Biol 2008; 182: 19–26.
Shire K, Ceccarelli DF, Avolio-Hunter TM, Frappier L. EBP2, a human protein that interacts with sequences of the Epstein-Barr virus nuclear antigen 1 important for plasmid maintenance. J Virol 1999; 73: 2587–2595.
Kapoor P, Lavoie BD, Frappier L . EBP2 plays a key role in Epstein-Barr virus mitotic segregation and is regulated by aurora family kinases. Mol Cell Biol 2005; 25: 4934–4945.
Shirai C, Takai T, Nariai M, Horigome C, Mizuta K . Ebp2p the yeast homolog of Epstein-Barr virus nuclear antigen 1-binding protein 2, interacts with factors of both the 60 S and the 40 s ribosomal subunit assembly. J Biol Chem 2004; 279: 25353–25358.
Tsujii R, Miyoshi K, Tsuno A, Matsui Y, Toh-e A, Miyakawa T et al. Ebp2p, yeast homologue of a human protein that interacts with Epstein-Barr virus nuclear antigen 1, is required for pre-rRNA processing and ribosomal subunit assembly. Genes Cell 2000; 5: 543–553.
Huber MD, Dworet JH, Shire K, Frappier L, McAlear MA . The budding yeast homolog of the human EBNA1-binding protein 2 (Ebp2p) is an essential nucleolar protein required for pre-rRNA processing. J Biol Chem 2000; 275: 28764–28773.
Shimoji K, Jakovljevic J, Tsuchihashi K, Umeki Y, Wan K, Kawasaki S et al. Ebp2 and Brx1 function cooperatively in 60S ribosomal subunit assembly in Saccharomyces cerevisiae. Nucleic Acids Res 2012; 40: 4574–4588.
Romanova L, Grand A, Zhang L, Rayner S, Katoku-Kikyo N, Kellner S et al. Critical role of nucleostemin in pre-rRNA processing. J Biol Chem 2009; 284: 4968–4977.
Nayyar VK, Shire K, Frappier L . Mitotic chromosome interactions of Epstein-Barr nuclear antigen 1 (EBNA1) and human EBNA1-binding protein 2 (EBP2). Journal of cell science 2009; 122 (Pt 23): 4341–4350.
Lee MC, Hsieh CH, Wei SC, Shen SC, Chen CN, Wu VC et al. Ectopic EBP2 expression enhances cyclin E1 expression and induces chromosome instability in HEK293 stable clones. BMB Rep 2008; 41: 716–721.
Reimers K, Antoine M, Zapatka M, Blecken V, Dickson C, Kiefer P . NoBPs a nuclear fibroblast growth factor 3 binding protein, is cell cycle regulated and promotes cell growth. Mol Cell Biol 2001; 21: 4996–5007.
Welcker M, Larimore EA, Frappier L, Clurman BE . Nucleolar targeting of the fbw7 ubiquitin ligase by a pseudosubstrate and glycogen synthase kinase 3. Mol Cell Biol 2011; 31: 1214–1224.
Liu N, Li H, Li S, Shen M, Xiao N, Chen Y et al. The Fbw7/human CDC4 tumor suppressor targets proproliferative factor KLF5 for ubiquitination and degradation through multiple phosphodegron motifs. J Biol Chem 2010; 285: 18858–18867.
Wang R, Hu H, Pan Y, Li Y, Ye T, Li C et al. RET fusions define a unique molecular and clinicopathologic subtype of non-small-cell lung cancer. J Clin Oncol 2012; 30: 4352–4359.
Leung AK, Trinkle-Mulcahy L, Lam YW, Andersen JS, Mann M, Lamond AI . NOPdb: Nucleolar Proteome Database. Nucleic Acids Res 2006; 34 (Database issue) D218–D220.
Ahmad Y, Boisvert FM, Gregor P, Cobley A, Lamond AI . NOPdb: Nucleolar Proteome Database—2008 update. Nucleic Acids Res 2009; 37 (Database issue): D181–D184.
Liao JM, Lu H . ChIP for identification of p53 responsive DNA promoters. Methods Mol Biol 2013; 962: 201–210.
Chang HY, Huang HC, Huang TC, Yang PC, Wang YC, Juan HF . Ectopic ATP synthase blockade suppresses lung adenocarcinoma growth by activating the unfolded protein response. Cancer Res 2012; 72: 4696–4706.
Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci USA 2004; 101: 9085–9090.
Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J 2004; 23: 2116–2125.
Blackwood EM, Eisenman RN . Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science 1991; 251: 1211–1217.
Chan JC, Hannan KM, Riddell K, Ng PY, Peck A, Lee RS et al. AKT promotes rRNA synthesis and cooperates with c-MYC to stimulate ribosome biogenesis in cancer. Sci Signal 2011; 4: ra56.
Poortinga G, Wall M, Sanij E, Siwicki K, Ellul J, Brown D et al. c-MYC coordinately regulates ribosomal gene chromatin remodeling and Pol I availability during granulocyte differentiation. Nucleic Acids Res 2011; 39: 3267–3281.
Gomez-Roman N, Felton-Edkins ZA, Kenneth NS, Goodfellow SJ, Athineos D, Zhang J et al. Activation by c-Myc of transcription by RNA polymerases I, II and III. Biochem Soc Symp 2006; 73: 141–154.
Acknowledgements
We thank Qi Deng, Dongmei Liu, and other members of the Wang lab for their assistances. This work was supported by grants from the National Basic Research Program of China (973 program 2010CB529704 and 2012CB910404), the National Natural Science Foundation of China (30800587, 30971521, and 31171338), and a grant from the Science and Technology Commission of Shanghai Municipality (11DZ2260300). PW is a scholar of the Shanghai Rising-Star Program from Science and Technology Commission of Shanghai Municipality (09QA1401900 and 13QH1401), the Program for New Century Excellent Talents in University (NCET-10-0387), and the Dawn Program of Shanghai Education Commission (11SG27).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Melino
Supplementary Information accompanies this paper on Cell Death and Disease website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Liao, P., Wang, W., Shen, M. et al. A positive feedback loop between EBP2 and c-Myc regulates rDNA transcription, cell proliferation, and tumorigenesis. Cell Death Dis 5, e1032 (2014). https://doi.org/10.1038/cddis.2013.536
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2013.536
Keywords
This article is cited by
-
lncRNA-PLACT1 sustains activation of NF-κB pathway through a positive feedback loop with IκBα/E2F1 axis in pancreatic cancer
Molecular Cancer (2020)
-
The long non-coding RNA FOXD2-AS1 promotes bladder cancer progression and recurrence through a positive feedback loop with Akt and E2F1
Cell Death & Disease (2018)
-
PTENβ is an alternatively translated isoform of PTEN that regulates rDNA transcription
Nature Communications (2017)
-
SCP1 regulates c-Myc stability and functions through dephosphorylating c-Myc Ser62
Oncogene (2016)
-
Nucleolus-derived mediators in oncogenic stress response and activation of p53-dependent pathways
Histochemistry and Cell Biology (2016)