Abstract
Mitochondrial morphology is dynamically regulated by the formation of small fragmented units or interconnected mitochondrial networks, and this dynamic morphological change is a pivotal process in normal mitochondrial function. In the present study, we identified a novel regulator responsible for the regulation of mitochondrial dynamics. An assay using CHANG liver cells stably expressing mitochondrial-targeted yellow fluorescent protein (mtYFP) and a group of siRNAs revealed that T-cell intracellular antigen protein-1 (TIA-1) affects mitochondrial morphology by enhancing mitochondrial fission. The function of TIA-1 in mitochondrial dynamics was investigated through various biological approaches and expression analysis in human specimen. Downregulation of TIA-1-enhanced mitochondrial elongation, whereas ectopic expression of TIA-1 resulted in mitochondria fragmentation. In addition, TIA-1 increased mitochondrial activity, including the rate of ATP synthesis and oxygen consumption. Further, we identified mitochondrial fission factor (MFF) as a direct target of TIA-1, and showed that TIA-1 promotes mitochondrial fragmentation by enhancing MFF translation. TIA-1 null cells had a decreased level of MFF and less mitochondrial Drp1, a critical factor for mitochondrial fragmentation, thereby enhancing mitochondrial elongation. Taken together, our results indicate that TIA-1 is a novel factor that facilitates mitochondrial dynamics by enhancing MFF expression and contributes to mitochondrial dysfunction.
Similar content being viewed by others
Main
Mitochondria continuously change their morphology by fusing or dividing in response to different cellular needs.1 Tight regulation of mitochondrial morphology is critical for the maintenance of mitochondrial structure and function, which ultimately affect cell fate.2, 3, 4 Disruption of dynamic balance is related to several physiological and pathological conditions such as aging, apoptosis, cancer, neurodegenerative diseases, and diabetes.5, 6, 7 Mitochondrial dynamics are governed by several core proteins. In mammals, mitofusin 1 (MFN1) and 2 (MFN2) are required for outer membrane fusion of mitochondria and optic atrophy protein 1 (OPA1) is involved in inner membrane fusion.8, 9, 10, 11 Dynamin-related protein 1 (DRP1), mitochondrial fission 1 (FIS1), mitochondrial dynamics 51 (MiD51) and MiD49 are core components of the mitochondrial fission machinery.12, 13, 14 Further, mitochondrial fission factor (MFF) is also a critical factor for mitochondrial fission by mediating recruitment of the DRP1 to mitochondria. MFF contains heptad repeats and a C-terminal transmembrane domain that is embedded in the outer membrane. Downregulation of MFF elongates the mitochondrial network.14, 15, 16
The rates of fusion and fission must be tightly regulated in order to preserve the correct balance required for the maintenance of mitochondrial morphology or to change the mitochondrial network in response to physiological needs.3 Several reports have shown that the expression of core proteins responsible for mitochondrial fusion or fission is controlled transcriptionally, post transcriptionally or post translationally.17, 18, 19 In particular, DRP1 activity is rapidly regulated by phosphorylation. Phosphorylation of serine616 in DRP1 by cyclin B1-cyclin-dependent kinase (cyclin B1-CDK1) or calcium–calmodulin-dependent kinase (CamK) enhances mitochondrial fission.20, 21 Phosphorylation of serine637 in DRP1 by protein kinase A (PKA) inhibits DRP1 function, whereas dephosphorylation by the calcium-sensitive protein phosphatase, calcineurin, enhances mitochondrial fission.22, 23 Moreover, DRP1 activity is also post translationally regulated by ubiquitin ligase membrane-associated RNIG-CH protein 5 (MARCH5) and by small ubiquitin-like modifier type 1 (SUMO1).24, 25, 26, 27, 28 micoRNA-499 (miR-499) is also involved in the regulation of DRP1 activity by targeting calcineurin at the post-transcriptional level.29 Hypoxia-inducible factor 1-alpha (HIF1α) promotes DRP1 activation and enhances mitochondrial fission.30, 31 Ubiquitination by MARCH5 or transcriptional control by PGC1β affects mitochondrial fusion through the regulation of MFN2.28, 32 miR-761 regulates the mitochondrial network by targeting MFF.33 Although several studies have been highlighted the significance of regulatory mechanisms governing the expression or activity of mitochondrial dynamics-regulating proteins such as DRP1 and MFN2, the specific regulators for the fusion–fission machinery and their roles during physiological or pathological conditions remain largely unknown.
T-cell-restricted intracellular antigen 1 (TIA-1) is an RNA-binding protein that functions as a posttranscriptional regulator of gene expression by binding to cis elements found in 5′- or 3′-untranslated regions (3′-UTRs) of selected mRNAs.34, 35, 36 TIA-1 has been reported to participate in translational repression in response to various stresses, as well as in the regulation of alternative splicing of target mRNAs.37, 38 It has been shown that mRNAs encoding tumor necrosis factor alpha (TNFα), cyclooxygenase 2 (COX2), and cytochrome C, and mRNAs containing U-rich elements or 5′-terminal oligopyrimidine tracts (5′TOP) are targets for TIA-1.35, 39, 40, 41, 42 Moreover, TIA-1 also functions in the stress granule (SGs) assembly in response to environmental stresses.43, 44
In the present study, we demonstrate that TIA-1 has a novel function in the regulation of mitochondrial dynamics. Downregulation of TIA-1 enhanced mitochondrial elongation, whereas ectopic expression of TIA-1 resulted in mitochondrial fragmentation. In addition, TIA-1 increased mitochondrial activity, including the rate of ATP synthesis and oxygen consumption. Further, we identified MFF as a direct target of TIA-1 and showed that TIA-1 promotes mitochondrial fragmentation by enhancing MFF translation. TIA-1 null cells had a decreased level of MFF and less mitochondrial Drp1, a critical factor for mitochondrial fragmentation, thereby enhancing mitochondrial elongation. Moreover, we observed that the relative expression of TIA-1 and MFF was higher in hepatocellular carcinoma (HCC). Taken together, our results suggest that TIA-1 facilitates mitochondrial fission by regulating MFF expression, and that the augmentation of TIA-1 expression may be responsible for mitochondrial dysfunction in HCC.
Results
TIA-1 is a novel regulator governing the morphological change of mitochondria
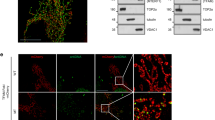
Several RBPs such as human antigen Hu, AUF1, or TIA-1, mediate various cellular processes through the regulation of RNA metabolism.45, 46, 47 To investigate the involvement of RBPs in the morphological changes in mitochondria, we examined mitochondrial morphology by following mitochondrial-targeted yellow fluorescent protein (mtYFP) in CHANG liver cells after transfection of siRNAs targeting various RBPs. Although the mitochondrial morphology was not affected by siRNA transfection, TIA-1 silencing resulted in significant changes inYFP-tagged mitochondria in CHANG liver cells (Supplementary Figure S1). We decided to further examine whether TIA-1 regulates mitochondrial dynamics. To this end, we evaluated the morphological changes in mitochondria in CHANG-mtYFP cells after transfection of siTIA-1 or HA-tagged TIA-1 plasmid, using appropriate controls. As shown in Figure 1, TIA-1 overexpression increased the number of cells showing fragmented mitochondria, whereas TIA-1 silencing increased the number of cells having elongated mitochondria (Figures 1a and b). We further investigated the mitochondrial morphology in TIA-1 null cells (TIA-1 −/− MEF cells) and control mouse embryonic fibroblast (MEF) cells (TIA-1 +/+ MEF cells). As shown in Figures 1c and d, mitochondria in TIA-1 null cells were significantly elongated compared with the control cells. Further analysis using transmission electron microscopy (TEM) revealed that TIA-1 null cells have more elongated forms of mitochondria (Figure 1e). These findings suggest that TIA-1 may act as a novel factor in the regulation of morphological changes in mitochondria.
TIA-1 mediates morphological changes in mitochondria. (a and b) CHANG liver cells expressing mtYFP were transiently transfected with siTIA-1 or TIA-1 plasmid, using proper controls. Forty-eight hours after transfection, YFP signals were observed under a fluorescence microscope. The percentage of cells with the indicated mitochondrial morphologies (intermediate, elongated, or fragmented forms) among 100 cells were analyzed in three independent experiments (top). TIA-1 levels were analyzed by western blotting using a TIA-1 antibody, and β-actin levels are shown as a loading control (bottom). (c and d) Mouse embryonic fibroblast cells lacking TIA-1 (TIA-1 −/−) and TIA+/+ cells were incubated with Mitotracker, and mitochondrial morphology was analyzed according to their relative length. TIA-1 levels in both MEF cells are shown by western blot analysis. (e) Representative transmission electron microscopic images of both MEF cells. The data represent the means±S.E.M. from three independent experiments. *P<0.05, **P<0.01, Bar, 20 μm
TIA-1 is necessary for mitochondrial function
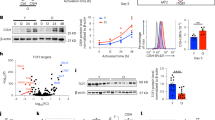
As dynamic changes in mitochondrial morphology are essential for their function, including mitochondrial respiration and/or ATP generation, we determined mitochondrial membrane potential by JC-1 staining and tetramethylrhodamine methyl ester (TMRM) fluorescence analysis following TIA-1 modulation in human HCC Hep3B cells. TIA-1 overexpression decrease, whereas TIA-1 downregulation increased, the portion of cells statined with JC-1 (Figure 2a). Also, TIA-1 overexpression downregulated, although TIA-1 downregulation resulted in an increase of TMRM fluorescence (Figure 2b). Next, we measured mitochondrial ATP level after TIA-1 overexpression or silencing. TIA-1 overexpression decreased, whereas TIA-1 downregulation increased, the mitochondrial ATP level (Figure 2c). We also investigated the impact of TIA-1 on cellular oxygen consumption rate (OCR) by using a Seahorse instrument. As shown in Figure 2d, TIA-1 overexpression impaired mitochondrial respiration and reduced basal OCR. Inversly, TIA-1 silencing resulted in an increase of basal OCR. Taken together, these results suggest that TIA-1 functions as a negative regulator in the maintenance of mitochondrial function.
TIA-1 mediates the regulation of mitochondrial function. After 48 h transfection of pTIA-1 or TIA-1 siRNA, Hep3B cells were stained with JC-1 (a) or TMEM (b) dyes. Relative fluorescence of JC-1 was determined according to manufacturer’s instruction. TMRM fluorescenct images were observed by conforcal microscope. (c) After transfection, cells were incubated with galactose-containing media for 90 min and mitochondrial ATP levels were determined as described in Materials and Methods. (d) Oxygen consumption rates were determined by Seahorse analysis using cells under basal condition and after sequential injection of an ATPase inhibitor (oligomycin), a mitochondrial uncoupler (FCCP), and an inhibitor of electron transport (Rotenone). Basal OCR was calculated from four different analysis. The data represent the mean±S.E.M. from at least three independent experiments. **P<0.01
TIA-1 binds to MFF mRNA 3′-UTR and enhances MFF translation
Since TIA-1 is one of the RBPs and affects target gene expression at their mRNA level by regulating alternative splicing, translation, and mRNA stability,35, 36, 38 we hypothesized that TIA-1 may be responsible for the expression of regulatory proteins that affect mitochondrial morphology, such as MFN1, MFN2, DRP1, OPA1, FIS1, MFF, and mitochondrial elongation factor 1 (MIEF1). To examine this possibility, we tested the association between TIA-1 and these mRNAs. Using a specific antibody against TIA-1, ribonucleoprotein (RNP) complexes containing TIA-1 were immunoprecipitated from Hep3B cell lysates, and associated mRNAs in IP complexes were further analyzed by RT-qPCR using the specific primer set listed in Supplementary Table 1. Although MFN1, MFN2, DRP1, OPA1, FIS1, and MIEF1 mRNA showed no significant interaction with TIA-1, MFF mRNA was highly enriched with TIA-1 containing the RNP complex (Figure 3a), indicating that TIA-1 interacts with MFF mRNA. To examine potential TIA-1 binding regions in MFF mRNA, pull down assays using biotin-labeled RNA probes were performed. Biotinylated segments of the MFF mRNA spanning the 5′-UTR and 3′-UTR (3U1, 3U2, 3U3, 3U1+2, and 3U) were synthesized (Figure 3b), and the RNP complexes containing TIA-1 and the biotinylated RNAs were isolated using streptavidin-coated beads and detected by western blot analysis using a TIA-1 antibody. As shown in Figure 3b (bottom), TIA-1 showed specific interactions with the 3U2, 3U3, 3U1+2, and 3U fragments of the MFF mRNA, but not with 5U or 3U1 or with the negative control, the 3′-UTR of GAPDH mRNA. In summary, TIA-1 associates with MFF mRNA through its 3′-UTR.
TIA-1 interacts with MFF mRNA. (a) TIA-1-associated mRNAs were isolated by RNP-IP using a TIA-1 antibody and analyzed by RT-qPCR. Seven mRNAs (MFN1, MFN2, DRP1, OPA1, FIS1, MFF, and MIEF1) were analyzed and GAPDH mRNA was used for normalization. The data represent the mean±S.E.M. from three independent experiments. (b) Schematic description of human MFF mRNA and biotinylated transcripts spanning the 5′-UTR (5U), coding region (CR), and 3′-UTRs (3U1, 3U2 and 3U3) (upper). Each biotinylated transcript was incubated with CHANG cell lysates and the interaction between TIA-1 and biotinylated transcripts was analyzed by western blot analysis using a TIA-1 antibody (lower). Data are representative of three independent experiments. *P<0.05
To evaluate whether TIA-1 modulates MFF expression, we investigated the mRNA and protein levels of MFF after TIA-1 overexpression and downregulation in Hep3B cells. As shown in Figure 4a, TIA-1 overexpression did not change MFF mRNA, whereas TIA-1 silencing slightly downregulated it. Although there was no significant changes in MFF mRNA, MFF protein level was positively affected by TIA-1; TIA-1 overexpression increased, whereas TIA-1 downregulation decreased MFF expression (Figure 4b). Cytochrome c level was negatively regulated by TIA-1 as described in previous report40 and it was shown as a positive control. Downregulation of MFF was also observed in TIA-1 null cells (Figure 4c), and TIA-1 overexpression in null cells resulted in an increase in MFF levels (Figure 4d). To further validate the regulation of MFF expression by TIA-1, we generated EGFP reporter plasmids containing the 3U2 fragment of MFF mRNA and examined EGFP expression after TIA-1 overexpression or silencing. As shown in Figure 4e, TIA-1 overexpression increased the expression of EGFP reporter containing 3U2 fragment of MFF mRNA and TIA-1 silencing decreased, but not the level of control EGFP, whereas TIA-1 silencing resulted in a decrease. Since MFF expression was affected by TIA-1 at the protein level rather than at the mRNA level, we evaluated the de novo synthesis of the EGFP reporter after TIA-1 overexpression. TIA-1 overexpression resulted in an increase in newly synthesized EGFP reporter containing the 3U2 fragment of MFF mRNA (Figure 4f). Altogether, these results indicate that TIA-1 positively regulates MFF expression by increasing its translation rather than affecting mRNA stability.
TIA-1 positively regulates MFF expression. (a and b) Following transfection of siTIA-1 or the TIA-1 plasmid, using proper controls, MFF mRNA and protein levels were analyzed by RT-qPCR (a) and western blotting (b). GAPDH mRNA and β-actin were used for normalization and loading control, respectively. Cytochrome c was used as a positive control. Data are representative of three independent experiments and values are expressed as the mean±S.E.M. (c) mRNA and protein levels in MEF TIA-1 +/+ and TIA-1 −/− cells (c) or MEF TIA-1 −/− cells following TIA-1 overexpression (d). (e) Schematic representation of EGFP reporters (upper). Hep3B cells expressing TIA-1 siRNAs or plasmids were sequentially transfected with EGFP reporters and EGFP levels were determined by western blot analysis (lower). (f) Nascent EGFP reporter production after TIA-1 overexpression. After transfection of the TIA-1 plasmid, cells were incubated with 35S-labeled methionine or cysteine for twenty minutes and EGFP proteins were immunoprecipitated using a GFP antibody and assessed by electrophoresis. Radioreactivity was analyzed using PharoseFX Plus. Data are representative of three independent experiments and values are expressed as the mean±S.E.M. *P<0.05
As a balance of fission and fusion events mediated by several regulatory proteins, we assessed whether TIA-1 affects other mitochondria-shaping proteins including DRP1, FIS1, MFN1/2, OPA1, and MIEF1 in Hep3B cells. TIA-1 overexpression or TIA-1 downregulation resulted in no significant changes in mRNA and protein levels of mitochondria-shaping proteins except MFF (Figures 5a and b). In addition, we did not observed differential expression of DRP1, MFN1/2, and OPA1 in TIA-1 −/− MEF cells (Figure 5c), which suggests that TIA-1 affects mitochondrial dynamics via MFF expression.
TIA-1 promotes MFF expression but not other mitochondria-shaping rpoteins. (a and b) Forty-eight-hour transfection of TIA-1 plasmid or siRNA in Hep3B cells, mRNA or proteins levels were analyzed by RT-qPCR or western blotting. GAPDH mRNA or β-actin was used for normalization. (c) Cell lysates from wile type MEF or TIA-1 −/− cells were prepared and relative protein expression was determined by western blotting. Graphs represent the mean±S.E.M. from three independent experiments. **P<0.01
TIA-1 competes with miR-27 to promote MFF expression
Our previous report indicates that microRNA-27 (miR-27) promotes mitochondrial elongation by reducing MFF expression.48 As we observed that TIA-1 displayed the opposite effect on the morphological changes in mitochondria (Figure 1), and the potential binding regions of miR-27 and TIA-1 were in close proximity, we speculated whether MFF expression is competitively regulated by TIA-1 and miR-27. To test this possibility, we investigated MFF expression after TIA-1 overexpression in cells transfected with a miR-27 mimic. As we observed previously, MFF levels were reduced by miR-27 transfection. However, the inhibitory effect of miR-27 was diminished by TIA-1 overexpression (Figure 6a). Further, in order to elucidate the manner in which TIA-1 and miR-27 affect MFF expression, we also investigated the association of TIA-1 with MFF mRNA in cells transfected with miR-27 following immunoprecipitation of TIA-1 containing RNP complexes. As shown in Figure 6b, miR-27 transfection resulted in a decrease in the association between TIA-1 and MFF mRNA, indicating that TIA-1 and miR-27 competitively regulate MFF expression.
TIA-1 alleviates miR-27 function in MFF expression. (a) Hep3B cells were co-transfected with TIA-1 plasmids and/or a miR-27 mimic. Forty-eight hours post transfection, MFF expression was assessed by western blotting using an MFF antibody. β-actin levels are shown as a loading control. (b) Following transfection of the miR-27 mimic, TIA-1-containing RNP complexes were isolated by RNP-IP using a TIA-1 antibody, and the relative MFF mRNA levels were determined by RT-qPCR. GAPDH mRNA was used for normalization. Data represent the mean±S.E.M. from three independent experiments. **P<0.01
The TIA-1/MFF axis promotes translocation of Drp1 to mitochondria
We observed a decrease in mitochondrial activity following TIA-1 overexpression (Figure 2) and positive regulation of MFF by TIA-1 (Figure 4). To investigate whether downregulation of mitochondrial membranepotential by TIA-1 overexpression is mediated via MFF regulation, we measured mitochondrial membrane potential in TIA-1-overexpressing cells after transfection with MFF siRNA (Figure 7a). As we observed previously, TIA-1 overexpression resulted in a decrease of mitochondrial membrane potential (lane 1 versus lane 3), whereas MFF silencing in TIA-1 overexpressing cells failed to reduce mitochondrial activity (lane 3 versus lane 4). These results indicate that TIA-1 regulates mitochondrial activity in an MFF-dependent manner.
The TIA-1/MFF axis promotes translocation of Drp1 to mitochondria. (a) CHANG cells were transiently co-transfected with siMFF and/or TIA-1 plasmids. Forty-eight hours post transfection, mitochondrial membrane potential was determined by the JC-1 staining assay or MFF levels were analyzed by western blotting using MFF antibody. Values represent the mean±S.E.M. from three independent experiments. (b) Subcellular distribution of Drp1 protein in TIA-1 +/+ and TIA-1 −/− cells. Mitochondrial fractions were isolated from both MEF cells and the relative MFF and Drp1 levels were assessed by western blot analysis. β-tubulin and Tom40 antibodies were used as markers for cytosolic and mitochondrial fractions, respectively. Data are representative from three independent experiments. **P<0.01
As it is known that MFF promotes mitochondrial fission by facilitating the translocation of Drp1 from the cytosol to mitochondria,15 we further investigated the relative levels of mitochondria-associated-Drp1 in TIA-1 null cells. Mitochondrial fractions were prepared by differential centrifugation from the lysates of both TIA-1 null cells and wild-type control cells, and Drp1 levels were assessed by western blotting (Figure 7b). Although there was no difference in total Drp1 level between TIA-1 null cells and control cells, we observed a reduction of Drp1 in the mitochondrial fraction of TIA-1 null cells, suggesting depletion of TIA-1 reduced mitochondrial translocation of Drp1 via the downregulation of MFF. Taken together, our findings indicate that the TIA-1/MFF axis mediates mitochondrial dynamics by modulating Drp1 translocation.
Discussion
Mitochondria execute pivotal roles in the regulation of energy metabolism, calcium signaling, and apoptosis, and dysfunction of mitochondria is related to the pathogenesis of several diseases.1, 3, 5, 49 Dynamic regulation of mitochondria is essential for the maintenance of cellular homeostasis and is governed by several key regulatory proteins such as Drp1, Mfn1/2, OPA1, FIS1, and MFF.1, 3, 17 In the present study, we demonstrated that an RNA-binding protein, TIA-1, functions as a novel factor affecting mitochondrial fission via MFF regulation. We showed that TIA-1 binds to MFF mRNA and promotes MFF expression, thereby enhancing mitochondrial fission. We further demonstrated that TIA-1 deficiency is related to enhanced mitochondrial activities including mitochondrial membrane potential, ATP synthesis, and OCRs. Our results suggest that the TIA-1/MFF axis has a novel role in the regulation of mitochondrial dynamics.
Although there are some controversial points, recent reports have demonstrated that mitochondrial dysfunction is mainly involved in the pathogenesis of several human diseases including neurodegenerative diseases and cancers, and that mitochondrial dynamics may offer potential therapeutic targets.3, 4, 6, 17, 50 In lung and breast cancers in particular, an increase in mitochondrial fission promotes cell cycle progression or migration.51, 52 Augmented expression of mitochondrial fission factors, such as Drp1, or reduced levels of mitochondrial fusion proteins are responsible for the imbalance of mitochondrial dynamics leading to mitochondrial dysfunction. To our knowledge, we present the first evidence that TIA-1 functions as pro-fission protein via promoting MFF expression at RNA level. Although we focused on the TIA-1/MFF axis in the regulation of mitochondrial dynamics in the present study, identification of other factors at the posttranscriptional level awaits further investigation in order to elucidate a novel regulatory mechanism governing mitochondrial dynamics.
TIA-1 is one of the RNA-binding proteins that regulates alternative splicing, translation, and turnover of certain mRNAs including TNFα and cytochrome c.14, 35, 39 In the present study, we identified MFF mRNA as a novel target of TIA-1 and demonstrated that TIA-1 enhances MFF expression by binding to its 3′-UTR (Figure 4). Despite previous reports showing TIA-1 as a translational suppressor, we observed that TIA-1 enhances MFF expression; TIA-1 overexpression increased MFF expression, while TIA-1 downregulation decreased it. As evidenced by our recent study, miR-27 targets MFF and contributes to the elongation of mitochondria,48 thus, we hypothesized that TIA-1 may compete with miR-27 to regulate MFF expression. As the TIA-1-binding region of MFF mRNA is in close proximity to the targeting sites of miR-27, and ectopic expression of miR-27 decreased the interaction between TIA-1 and MFF mRNA (Figure 6b), we suggest the competitive regulation of MFF expression by TIA-1 and miR-27. Although several studies have shown that competitive or cooperative interplay among regulatory factors including RBPs and miRNAs is essential for the fine-tuning of gene expression,53, 54, 55, 56 the detailed mechanism of such regulation has not yet been fully elucidated. We believe further studies will enable a better understanding of the manner in which regulatory factors interplay to contribute to the dynamic regulation of gene expression.
In addition, there are some discrepancies among previous reports regarding the role of TIA-1 in cell proliferation; depletion of TIA-1 in human cells (FT293T, HeLa and Hep3B) resulted in an increase in cell proliferation,58, 59, 60 whereas TIA-1-deficient mouse embryonic fibroblasts showed a decrease in cell growth.61 Moreover, it appeares that TIA-1 expression is differentially regulated in different types of cancer; TIA-1 is upregulated in HCC,57 while it is reduced in small-cell lung carcinoma (SCLC).60 Although we still do not know exactly why TIA-1 displays differential activity in the regulation of cell proliferation, it may be a result of a difference in species, cell types, and origin of cancers. Consequently, it seems difficult to conclude whether TIA-1 functions as a tumor suppressor or an oncogene without comprehensive understanding of the TIA-1 working mechanism, thus, this deserves further investigation.
We believe that morphology and function of mitochondria influence each other; therefore, it is difficult to determine what comes first, morphological changes or dysfunction. Several lines of evidence indicate that a correlation between mitochondrial morphology and their functional state is not one-to-one correlation. For example, Rotenone treatment increased mitochondrial fragmentation, which suggests mitochondrial dysfunction leads to morphological changes (fission).62 Conversely, loss of mitochondrial fusion proteins such as MFN1/2 and OPA1 decreased mitochondrial membrane potential.63, 64, 65 In this study, we demonstrated that TIA-1 overexpression also resulted in a decrease of basal respiration of mitochondrial. Although we showed TIA-1 promotes mitochondrial fission via enhancing MFF expression, we still do not know whether TIA-1 cause mitochondrial dysfunction in MFF-independent manner or not. We believe further studies will be helpful to determine what comes first by TIA-1, misshape or dysfunction.
In conclusion, our study suggests that TIA-1 is a novel pro-fission protein by enhancing MFF expression, thereby results in mitochondrial dysfunction. Further studies on differential expression of TIA-1 or MFF and their molecular mechanisms may provide additional signicicance of the TIA-1/MFF axis in the pathogenesis of several diseases including cancer and neurodegenerative diseases.
Materials and Methods
Cell culture, transfection, plasmid, and small interfering RNAs
Human CHANG liver cells stably overexpressing mitochondrial targeted YFP (mtYFP) were cultured in Dulbecco’s modified essential medium (Hyclone, Logan, UT, USA), supplemented with 10% fetal bovine serum and antibiotics.66 TIA-1-deficient mouse embryonic fibroblasts (MEF TIA-1 −/−) and HA-tagged TIA-1 plasmid were great gifts from Dr. Paul Anderson (Harvard Medical School, Boston, MA, USA).39 Enhanced green fluorescent protein (EGFP) reporters were cloned by inserting 3′-UTR fragments from the MFF mRNA (1751-2040, 290 nt) into pEGFP-C1 (BD Bioscience, Franklin lake, NJ, USA). Small interfering RNAs (siRNAs; control siRNA (siCtrl) and TIA-1 siRNA (siTIA-1); Genolution Pharmarceuticals, Seoul, Korea) or plasmids were transiently transfected using Lipofectamin 2000 (Invitrogen, Calsbad, CA, USA).
Western blot analysis
Whole-cell lysates were prepared using RIPA buffer containing 10 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% NP-40, 1 mM EDTA, 0.1% SDS, and 1 × protease inhibitor cocktail (Roche, Basel, Switzerland), separated by SDS polyacrylamide gel electrophoresis, and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA). The membranes were incubated with primary antibodies against MFF (Abcam, Cambridge, MA, USA), TIA-1 (Santa Cruz Biotechnology, Dallas, TX, USA), EGFP (Santa Cruz Biotech), Drp-1 (BD Bioscience) or β-actin (Abcam) then further incubated with the appropriate secondary antibodies conjugated with horseradish peroxidase (Santa Cruz Biotechnology). The signals were detected using enhanced luminescence (Bio-Rad, Hercules, CA, USA).
RNA analysis and ribonucleoprotein complex-immunoprecipitation
Total RNA was prepared from whole cells using Trizol (Invitrogen). After reverse transcription (RT) using random hexamers and reverse transcriptase (Toyobo, Osaka, Japan), the mRNA abundance was assessed by reverse transcription quantitative PCR (RT-qPCR) analysis using the SYBR green PCR master mix (Kapa Biosystems, Wilmington, MA, USA) and gene-specific primer sets (Supplementary Table 1). RT-qPCR reaction was performed using StepOne Plus system (Life Technologies, Calsbad, CA, USA).
Ribonucleoprotein complex-immunoprecipitation (RNP-IP) reaction was done using anti-TIA-1 primary antibodies or control IgG (Santa Cruz Biotechnology).67 In brief, RNP complexes were immunoprecipitated using TIA-1 or control IgG antibodies and incubated with DNase I and proteinase K; RNA in the immunoprecipitation (IP) samples was isolated and further analyzed by RT-qPCR using the primers listed (Supplementary Table 1).
Biotin pulldown analysis
To synthesize biotinylated transcripts, PCR fragments were prepared using forward primers that contained the T7 RNA polymerase promoter sequence (T7, 5′-CCAAGCTTCTAATACGACTCACTATAGGGAGA-3′). The primers used to prepare biotinylated transcripts spanning the MFF mRNA (NM_001277061.1) are listed in Supplementary Table 1. After purification of the PCR products, biotinylated transcripts were synthesized using the MaxiScript T7 kit (Ambion, Waltham, MA, USA) and biotin-CTP (Enzo Life Sciences, Farmingdale, NY, USA). Whole-cell lysates (200 μg per sample) were incubated with 1 μg of purified biotinylated transcripts for 30 min at room temperature, and then complexes were isolated using streptavidin-coupled Dynabeads (Invitrogen). Proteins present in the pulldown material were studied by western blot analysis as described in Bach et al.63
Fluorescence microscopy
Mitochondrial morphology was analyzed by tracing YFP signals from CHANG liver cells stably expressing mtYFP under the Axiovert 200M microscope (Carl Zeiss, Jena, Germany). For staining of mitochondria, cells were incubated in serum free medium supplemented with 100 nM MitoTracker Red CMXRos (Invitrogen) for 30 min. Morphological changes of mitochondria were analyzed under the Axiovert 200M microscope (Carl Zeiss) and images were acquired using an Axiovertcam mRM camera (Carl Zeiss).
Mitochondrial activity assay
The mitochondrial membrane potential was measured using the JC1 Mitochondrial Membrane Potential Assay Kit (Abcam) or MitoPT TMRM Assay Kit (Immunochemistry Technologies LLC, Bloomington, MN, USA). For JC-1 staining, cells were incubated with tetraethyl benzimidazoly carbocyanine iodide (JC-1) staining solution (Abcam) for 10 min at 37 °C in the dark, and the fluorescence was measured at 535 nm (excitation)/590 nm (emission) using a Victor3 fluorescent plate reader (Perkin Elmer Life Sciences, Waltham, MA, USA). For TMRM staining, cells were incubated with 200 nM tetramethylrhodamine methyl ester (TMRM) staining solution (Immunochemistry Technologies LLC) for 20 min and the fluorescence was measured at 510–540 nm (excitation)/570–620 nm (emission) using a LSM 510 Meta Confocal microscope (Carl Zeiss). Mitochondrial ATP level determined by performing the Mitochondrial ToxGlo assay according to the manufacturer’s procedure (Promega, Madison, WI, USA). Briefly, cells then incubated with galactose-containing media at 37 °C for 90 min and then further incubated with ATP Detection reagent. Luminescence was measured using a Victor3 plate reader (Perkin Elmer).
Subcellular fractionation
Cells were incubated with hypotonic buffer containing 20 mM HEPES (pH 7.3), 1.5 mM MgCl2, and 10 mM KCl on ice and lysed using 8% (w/v) CHAPS. Cell lysates were further incubated with hypertonic buffer containing 20 mM HEPES (pH 7.3), 420 mM mannitol, 140 mM sucrose, and 2 mM EGTA and centrifuged at 700 × g for 10 min. Supernatants were further centrifuged at 12 000 × g for 15 min.
Nascent translation assay
Nascent translation of EGFP was studied by incubating TIA-1 −/− MEF cells with 1 mCi L-[35S]methionine and L-[35S]cysteine (Perkin Elmer) per 35 mm plate for 15 min. Immunoprecipitation reactions were carried out using mouse IgG, or anti-GFP (Santa Cruz Biotechnology), and IP products were separated, transferred, and visualized using a PharoseFX Plus system (Bio-Rad) analyzed using Quantity One software (Bio-Rad).
Electron microscopy
For TEM observation, cells were fixed with 1% osmium tetroxide and embedded using Epon 812. Ultrathin sections were observed with a transmission electron microscope (JEM 1010, Tokyo, Japan).
Analysis of oxygen consumption
A Seahorse FX24 Extracellular Flux Analyzer (Seahorse Bioscience, North Billerica, MA, USA) was used to measure OCR according to the manufacturer’s instruction. Basal OCR was measured for 3 min every 8 min for four points, followed by sequential injection of olygomycin (3 μ M), FCCP (1 μ M), and antimycin A (1 μ M). Each treatment was measured for 3 min every 8 min for three points. The number of cells (1 × 103) was used for data normalization.
Abbreviations
- TIA-1:
-
T-cell-restricted intracellular antigen 1
- MFF:
-
Mitochondrial fission factor
- mtYFP:
-
yellow fluorescence protein targeting mitochondria
- 3′-UTR:
-
3′-untranslated region
- HCC:
-
Hepatocellular carcinoma
- DRP1:
-
Dynamin-related protein 1
- MEF:
-
mouse embryonic fibroblast
- OCR:
-
Oxygen consumption rate
- miR-27:
-
microRNA-27
- RNP:
-
Ribonucleoprotein
References
Campello S, Scorrano L . Mitochondrial shape changes: orchestrating cell pathophysiology. EMBO Rep 2010; 11: 678–684.
Hoppins S, Lackner L, Nunnari J . The machines that divide and fuse mitochondria. Annu Rev Biochem 2007; 76: 751–780.
Westermann B . Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol 2010; 11: 872–884.
Liesa M, Palacin M, Zorzano A . Mitochondrial dynamics in mammalian health and disease. Physiol Rev 2009; 89: 799–845.
Chan DC . Mitochondria: dynamic organelles in disease, aging, and development. Cell 2006; 125: 1241–1252.
Grandemange S, Herzig S, Martinou JC . Mitochondrial dynamics and cancer. Semin Cancer Biol 2009; 19: 50–56.
Suen DF, Norris KL, Youle RJ . Mitochondrial dynamics and apoptosis. Genes Dev 2008; 22: 1577–1590.
Santel A, Fuller MT . Control of mitochondrial morphology by a human mitofusin. J Cell Sci 2001; 114: 867–874.
Cipolat S, Martins de Brito O, Dal Zilio B, Scorrano L . OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc Natl Acad Sci USA 2004; 101: 15927–15932.
Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC . Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol 2003; 160: 189–200.
Meeusen S, McCaffery JM, Nunnari J . Mitochondrial fusion intermediates revealed in vitro . Science 2004; 305: 1747–1752.
Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM . A human dynamin-related protein controls the distribution of mitochondria. J Cell Biol 1998; 143: 351–358.
Zhang Y, Chan DC . Structural basis for recruitment of mitochondrial fission complexes by Fis1. Proc Natl Acad Sci USA 2007; 104: 18526–18530.
Gandre-Babbe S, van der Bliek AM . The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell 2008; 19: 2402–2412.
Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ et al. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol 2010; 191: 1141–1158.
Loson OC, Song Z, Chen H, Chan DC . Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol Biol Cell 2013; 24: 659–667.
Archer SL . Mitochondrial dynamics—mitochondrial fission and fusion in human diseases. N Engl J Med 2013; 369: 2236–2251.
Chauhan A, Vera J, Wolkenhauer O . The systems biology of mitochondrial fission and fusion and implications for disease and aging. Biogerontology 2013; 15: 1–12.
Palmer CS, Osellame LD, Stojanovski D, Ryan MT . The regulation of mitochondrial morphology: intricate mechanisms and dynamic machinery. Cell Signal 2011; 23: 1534–1545.
Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K . Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem 2007; 282: 11521–11529.
Han XJ, Lu YF, Li SA, Kaitsuka T, Sato Y, Tomizawa K et al. CaM kinase I alpha-induced phosphorylation of Drp1 regulates mitochondrial morphology. J Cell Biol 2008; 182: 573–585.
Cribbs JT, Strack S . Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep 2007; 8: 939–944.
Cereghetti GM, Stangherlin A, Martins de Brito O, Chang CR, Blackstone C, Bernardi P et al. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc Natl Acad Sci USA 2008; 105: 15803–15808.
Zunino R, Schauss A, Rippstein P, Andrade-Navarro M, McBride HM . The SUMO protease SENP5 is required to maintain mitochondrial morphology and function. J Cell Sci 2007; 120: 1178–1188.
Braschi E, Zunino R, McBride HM . MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep 2009; 10: 748–754.
Park YY, Lee S, Karbowski M, Neutzner A, Youle RJ, Cho H . Loss of MARCH5 mitochondrial E3 ubiquitin ligase induces cellular senescence through dynamin-related protein 1 and mitofusin 1. J Cell Sci 2010; 123: 619–626.
Karbowski M, Neutzner A, Youle RJ . The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J Cell Biol 2007; 178: 71–84.
Nakamura N, Kimura Y, Tokuda M, Honda S, Hirose S . MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep 2006; 7: 1019–1022.
Wang JX, Jiao JQ, Li Q, Long B, Wang K, Liu JP et al. miR-499 regulates mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nat Med 2011; 17: 71–78.
Marsboom G, Toth PT, Ryan JJ, Hong Z, Wu X, Fang YH et al. Dynamin-related protein 1-mediated mitochondrial mitotic fission permits hyperproliferation of vascular smooth muscle cells and offers a novel therapeutic target in pulmonary hypertension. Circ Res 2012; 110: 1484–1497.
Semenza GL . Oxygen sensing, homeostasis, and disease. N Engl J Med 2011; 365: 537–547.
Liesa M, Borda-d'Agua B, Medina-Gomez G, Lelliott CJ, Paz JC, Rojo M et al. Mitochondrial fusion is increased by the nuclear coactivator PGC-1beta. PLoS ONE 2008; 3: e3613.
Long B, Wang K, Li N, Murtaza I, Xiao JY, Fan YY et al. miR-761 regulates the mitochondrial network by targeting mitochondrial fission factor. Free Radic Biol Med 2013; 65: 371–379.
Anderson P, Nagler-Anderson C, O'Brien C, Levine H, Watkins S, Slayter HS et al. A monoclonal antibody reactive with a 15-kDa cytoplasmic granule-associated protein defines a subpopulation of CD8+ T lymphocytes. J Immunol 1990; 144: 574–582.
Lopez de Silanes I, Galban S, Martindale JL, Yang X, Mazan-Mamczarz K, Indig FE et al. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol Cell Biol 2005; 25: 9520–9531.
Stoecklin G, Anderson P . Posttranscriptional mechanisms regulating the inflammatory response. Adv Immunol 2006; 89: 1–37.
Forch P, Puig O, Kedersha N, Martinez C, Granneman S, Seraphin B et al. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol Cell 2000; 6: 1089–1098.
Forch P, Valcarcel J . Molecular mechanisms of gene expression regulation by the apoptosis-promoting protein TIA-1. Apoptosis 2001; 6: 463–468.
Piecyk M, Wax S, Beck AR, Kedersha N, Gupta M, Maritim B et al. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J 2000; 19: 4154–4163.
Kawai T, Lal A, Yang X, Galban S, Mazan-Mamczarz K, Gorospe M . Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol Cell Biol 2006; 26: 3295–3307.
Dixon DA, Balch GC, Kedersha N, Anderson P, Zimmerman GA, Beauchamp RD et al. Regulation of cyclooxygenase-2 expression by the translational silencer TIA-1. J Exp Med 2003; 198: 475–481.
Damgaard CK, Lykke-Andersen J . Translational coregulation of 5'TOP mRNAs by TIA-1 and TIAR. Genes Dev 2011; 25: 2057–2068.
Kedersha NL, Gupta M, Li W, Miller I, Anderson P . RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol 1999; 147: 1431–1442.
Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM et al. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 2004; 15: 5383–5398.
Moore MJ . From birth to death: the complex lives of eukaryotic mRNAs. Science 2005; 309: 1514–1518.
Pullmann R Jr., Kim HH, Abdelmohsen K, Lal A, Martindale JL, Yang X et al. Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs. Mol Cell Biol 2007; 27: 6265–6278.
Glisovic T, Bachorik JL, Yong J, Dreyfuss G . RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 2008; 582: 1977–1986.
Tak H, Kim J, Jayabalan AK, Lee H, Kang H, Cho DH et al. miR-27 regulates mitochondrial networks by directly targeting the mitochondrial fission factor. Exp Mol Med 2014; 46: e123.
McBride HM, Neuspiel M, Wasiak S . Mitochondria: more than just a powerhouse. Curr Biol 2006; 16: R551–R560.
Cho DH, Nakamura T, Lipton SA . Mitochondrial dynamics in cell death and neurodegeneration. Cell Mol Life Sci 2010; 67: 3435–3447.
Rehman J, Zhang HJ, Toth PT, Zhang Y, Marsboom G, Hong Z et al. Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer. FASEB J 2012; 26: 2175–2186.
Zhao J, Zhang J, Yu M, Xie Y, Huang Y, Wolff DW et al. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene 2013; 32: 4814–4824.
Tominaga K, Srikantan S, Lee EK, Subaran SS, Martindale JL, Abdelmohsen K et al. Competitive regulation of nucleolin expression by HuR and miR-494. Mol Cell Biol 2011; 31: 4219–4231.
Zhuang R, Rao JN, Zou T, Liu L, Xiao L, Cao S et al. miR-195 competes with HuR to modulate stim1 mRNA stability and regulate cell migration. Nucleic Acids Res 2013; 41: 7905–7919.
Lee EK, Kim HH, Kuwano Y, Abdelmohsen K, Srikantan S, Subaran SS et al. hnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies. Nat Struct Mol Biol 2010; 17: 732–739.
Yu TX, Rao JN, Zou T, Liu L, Xiao L, Ouyang M et al. Competitive binding of CUGBP1 and HuR to occludin mRNA controls its translation and modulates epithelial barrier function. Mol Biol Cell 2013; 24: 85–99.
Regel I, Eichenmuller M, Joppien S, Liebl J, Haberle B, Muller-Hocker J et al. IGFBP3 impedes aggressive growth of pediatric liver cancer and is epigenetically silenced in vascular invasive and metastatic tumors. Mol Cancer 2012; 11: 9.
Izquierdo JM, Alcalde J, Carrascoso I, Reyes R, Ludena MD . Knockdown of T-cell intracellular antigens triggers cell proliferation, invasion and tumour growth. Biochem J 2011; 435: 337–344.
Reyes R, Alcalde J, Izquierdo JM . Depletion of T-cell intracellular antigen proteins promotes cell proliferation. Genome Biol 2009; 10: R87.
Sanchez-Jimenez C, Ludena MD, Izquierdo JM . T-cell intracellular antigens function as tumor suppressor genes. Cell Death Dis 2015; 6: e1669.
Sanchez-Jimenez C, Izquierdo JM . T-cell intracellular antigen (TIA)-proteins deficiency in murine embryonic fibroblasts alters cell cycle progression and induces autophagy. PLoS ONE 2013; 8: e75127.
Benard G, Bellance N, James D, Parrone P, Fernandez H, Letellier T et al. Mitochondrial bioenergetics and structural network organization. J Cell Sci 2007; 120 (Pt 5): 838–848.
Bach D, Pich S, Soriano FX, Vega N, Baumgartner B, Oriola J et al. Mitofusin-2 determines mitochondrial network architecture and mitochondrial metabolism. A novel regulatory mechanism altered in obesity. J Biol Chem 2003; 278 (19): 17190–17197.
Chen H, Chomyn A, Chan DC . Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem 2005; 280 (28): 26185–26192.
Sebastian D, Hernandez-Alvarez MI, Segales J, Sorianello E, Munoz JP, Sala D et al. Mitofusin 2 (Mfn2) links mitochondrial and endoplasmic reticulum function with insulin signaling and is essential for normal glucose homeostasis. Proc Natl Acad Sci USA 2012; 109 (14): 5523–5528.
Yoon YS, Yoon DS, Lim IK, Yoon SH, Chung HY, Rojo M et al. Formation of elongated giant mitochondria in DFO-induced cellular senescence: involvement of enhanced fusion process through modulation of Fis1. J Cell Physiol 2006; 209: 468–480.
Lee EK, Kim W, Tominaga K, Martindale JL, Yang X, Subaran SS et al. RNA-binding protein HuD controls insulin translation. Mol Cell 2012; 45: 826–835.
Acknowledgements
This work is supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (2014R1A2A1A11053431, 2012M3A9D1054517, and 2012R1A5A2047939).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by L Scorrano
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Tak, H., Eun, J., Kim, J. et al. T-cell-restricted intracellular antigen 1 facilitates mitochondrial fragmentation by enhancing the expression of mitochondrial fission factor. Cell Death Differ 24, 49–58 (2017). https://doi.org/10.1038/cdd.2016.90
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2016.90