Abstract
Glioblastomas are highly aggressive brain tumours and are characterised by substantial cellular heterogeneity within a single tumour. A sub-population of glioblastoma stem-like cells (GSCs) that shares properties with neural precursor cells has been described, exhibiting resistance to therapy and therefore being considered responsible for the high recurrence rate in glioblastoma. To elucidate the underlying cellular processes we investigated the role of phosphatases in the GSC phenotype, using an in vitro phosphatome-wide RNA interference screen. We identified a set of genes, the knockdown of which induces a significant decrease in the glioma stem cell marker CD133, indicating a role in the glioblastoma stem-like phenotype. Among these genes, the ecto-nucleotidase ENPP1 (ectonucleotide pyrophosphatase/phosphodiesterase 1) was found to be highly expressed in GSCs compared with normal brain and neural stem cells. Knockdown of ENPP1 in cultured GSCs resulted in an overall downregulation of stem cell-associated genes, induction of differentiation into astrocytic cell lineage, impairment of sphere formation, in addition to increased cell death, accumulation of cells in G1/G0 cell cycle phase and sensitisation to chemotherapeutic treatment. Genome-wide gene expression analysis and nucleoside and nucleotide profiling revealed that knockdown of ENPP1 affects purine and pyrimidine metabolism, suggesting a link between ENPP1 expression and a balanced nucleoside–nucleotide pool in GSCs. The phenotypic changes in E-NPP1-deficient GSCs are assumed to be a consequence of decreased transcriptional function of E2F1. Together, these results reveal that E-NPP1, by acting upstream of E2F1, is indispensable for the maintenance of GSCs in vitro and hence required to keep GSCs in an undifferentiated, proliferative state.
Similar content being viewed by others
Main
Glioblastoma, classified by the WHO (World Health Organization) as grade IV astrocytoma,1 is the most common primary malignant brain tumour in adults. Despite progress in surgical resection, radiation and chemotherapy, glioblastoma remains a deadly disease with a median survival time of about 1 year.2 One particular therapeutic challenge for glioblastoma treatment is posed by their remarkable intratumoural cellular heterogeneity. Several studies indicate the existence of a highly tumourigenic, sub-population of cancer cells with stem-like characteristics.3, 4, 5 There is substantial evidence that these so-called cancer stem cells have inherent chemotherapy and radiation resistance.6, 7 These findings suggest that cancer cells harbouring stem cell-like characteristics are responsible for ineffective therapy, explaining high recurrence rates despite significant reduction in tumour volume. Glioblastoma stem-like cells (GSCs) share properties with neural precursor cells, such as a capacity for self-renewal, differentiation and maintained proliferation, as well as stem cell marker expression.4, 5, 8, 9 GSCs were originally defined by expression of CD133 (Prominin-1) and its extracellular AC133 epitope.4 Although recent studies have shown that GSCs are not entirely restricted to the CD133-positive cell population,10, 11 there is a large body of evidence linking enhanced CD133 expression to poor prognosis in glioblastoma.12, 13 The role of CD133-positive GSCs in tumourigenicity is further strengthened by a recent report, demonstrating that CD133 has an essential role in the self-renewal and tumourigenic potential of GSCs.14 In the past years, several studies have been conducted to investigate molecular mechanisms that regulate the maintenance of GSCs for a better understanding of tumour biology. A kinome-wide RNA interference (RNAi) screen reported several kinases that regulate the balance of GSC self-renewal and differentiation.15 In particular, the adaptor protein TRRAP (transformation/transcription domain-associated protein) was shown to have an important role in maintaining a tumourigenic, stem cell-like state. These findings underline the importance of phosphorylation processes as a regulator of stem cell relevant pathways. In the study presented here, we investigated the role of phosphatases in the GSC phenotype. For this purpose, we performed a phosphatome-wide RNAi screen targeting 177 human phosphatases with the read-out based on the CD133 stem cell marker level upon knockdown. Twelve phosphatases were identified, which positively regulate the CD133 level of GSCs in vitro, indicating that these genes are involved in the GSC phenotype. We functionally characterised ENPP1 that showed a cancer-specific expression, and showed that it is an important regulator of the glioblastoma stem-like phenotype.
Results
A phosphatome-wide RNAi screen identifies genes important for the glioblastoma stem-like phenotype
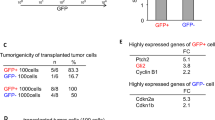
To identify phosphatases relevant for the glioblastoma stem-like phenotype we carried out a lentiviral-mediated loss-of-function screen using the stem cell marker CD133 (Prominin-1) as read-out. GSCs were grown in 96-well plates in stem cell medium and transduced with a lentiviral shRNA library targeting 177 phosphatases (an average of 4.1 shRNAs per gene). GSCs transduced with lentiviral particles containing GFP were used as transduction efficiency control and only plates with an efficiency of at least 70% were considered. Six days after lentivirus-mediated knockdown, GSCs were analysed for their CD133 level by flow cytometry, staining the AC133 epitope (CD133/1). As positive control, we used TRRAP-shRNA, which was previously identified as a regulator of GSC differentiation.15 GSCs transduced with TRRAP-shRNA (TRRAP-deficient cells) showed a decreased CD133/1 level compared with non-target shRNA (shNT) transduced GSCs (non-target control cells) expressed as median fluorescence intensity (MFI) above the isotype control (normalised MFI (nMFI)) (Figures 1a and b). Efficient knockdown was confirmed by quantitative real-time PCR (qRT-PCR) (Supplementary Figure S1A). Each plate harboured three wells with non-target control cells and two wells with TRRAP-deficient cells. To account for technical variability, data were normalised to the mean of negative (non-target control cells; CD133 level set to 1) and positive (TRRAP-deficient cells; CD133 level set to 0) controls of the respective plate. Untreated GSCs (Mock; one well per plate) served as further negative control, whereas GSCs treated with 10 μM all-trans retinoic acid (ATRA; two wells per plate), previously shown to induce differentiation of GSCs and thus to reduce the CD133 level,16 was used as a second positive control. As depicted in Supplementary Figure S1B, negative and positive controls on the screening plates (n=20) exhibit only minimal variability of CD133/1 level, underlining the high quality of the screen. Genes were considered potential hits if the CD133 levels were significantly decreased (false discovery rate (FDR)<0.05) compared with non-target control cells (Figure 1c). This led to the identification of 12 phosphatases potentially involved in the stem-like phenotype of GSCs (Figures 1c and d). Six of the twelve candidate genes (PPEF1, ENPP1, PTPN9, PPP4C, PPP2R2A and DUSP5) were found to be highly expressed in astrocytic gliomas compared with adult normal brain and/or to be essential for stem cell maintenance (Supplementary Figure S2A)17, 18, 19, 20, 21, 22 and hence selected for validation studies. Decreased CD133 level 6 days post-transduction was confirmed in NCH421k (GSCs used in the primary screen) and in additional GSCs (NCH441, NCH644) in two independent experiments using the two shRNAs that showed the strongest CD133 reduction in the primary screen (Figure 1e; Supplementary Figure S2B). Efficient downregulation of the target gene expression was verified by qRT-PCR (Supplementary Figure S2C).
Phosphatome-wide RNAi screen identifies genes important for the glioblastoma stem-like phenotype. (a) CD133/1 FACS analysis of NCH421k cells transduced with shNT and TRRAP-shRNA (shTRRAP), which served as negative and positive control for the screen, respectively. The red dotted line indicates the MFI of the respective histogram. (b) Quantification of FACS results shown in (a) using the nMFI. Mean values and S.D. are shown (*P<0.05, n=3). (c) Bar chart of the screen results. The −log10 P-values for each phosphatase is shown. The genes whose knockdown significantly decreased CD133/1 level in comparison with cells transduced with shNT were considered as positive hits. The red line indicates the threshold for significance, which was set to a FDR<0.05. (d) Heat map showing the candidate genes of the screen and their decreased level of CD133/1 after knockdown using 3–5 shRNAs per gene in comparison with non-target control. Data are presented as normalised to shNT (CD133 level set to 1) and shTRRAP (CD133 level set to 0). NA: not available. (e) Verification of reduced CD133/1 level upon candidate gene knockdown in three different GSC lines (NCH421k, NCH441 and NCH644). Two independent experiments and the corresponding mean of shRNA1 are shown. Data are normalised to shNT (CD133 level set to 1) and shTRRAP (CD133 level set to 0)
Elevated expression of ENPP1 in GSCs compared with normal brain
From the genes potentially involved in the stem-like phenotype of GSCs, we selected ENPP1 (ectonucleotide pyrophosphatase/phosphodiesterase 1) for further studies as it was shown to be strongly expressed in grade IV glioblastoma tumours compared with low-grade astrocytic tumours and normal adult brain (Aerts et al.17 Supplementary Figure S2A). To ascertain the expression of ENPP1 within the GSC population, we performed qRT-PCR analysis in 12 patient-derived GSCs, including 5 short-term cultured GSCs (⩽6 passages) and 1 adult human normal brain sample. As depicted in Figure 2a, GSCs showed an elevated ENPP1 mRNA expression when compared with adult normal brain extract. This result was confirmed at protein level in three GSC lines (NCH421k, NCH441 and NCH644) (Figure 2b). Notably, PTPN9, PPP4C and PPP2R2A were also found to be higher-expressed in GSCs compared with adult normal brain extract (Supplementary Figure S2D). Publicly available microarray data of ENPP1 mRNA expression in adherent GSCs compared with fetal neural stem cells (fNSCs)23 further confirmed tumour specific expression (P=0.03; Figure 2c).
Elevated E-NPP1 expression in GSCs compared with adult normal brain samples. (a) qRT-PCR analysis of ENPP1 mRNA expression in GSC lines derived from 12 individual patients’ tumours. Data are normalised to housekeeping genes and to a pool of normal brain samples. (b) Western blot analysis showing E-NPP1 protein expression in three different GSC lines (NCH421k, NCH441 and NCH644) and in adult normal brain sample. α-tubulin protein expression was used as loading control. (c) In silico analysis using publicly available microarray data, showing increased ENPP1 mRNA expression in adherent GSCs (n=6) compared with fNSCs (n=5, P=0.03, GSE15209)
ENPP1 knockdown induces differentiation of GSCs
To assess the functional role of E-NPP1 in GSCs, NCH421k and NCH441 cells were transduced with shNT, as well as two independent shRNAs targeting ENPP1 (ENPP1-shRNA1 and ENPP1-shRNA3), and efficient protein knockdown was confirmed by western blot analyses (Figure 3a, Supplementary Figure S3A). To verify our screening results, two independent CD133 epitopes, AC133 (CD133/1) and AC141 (CD133/2), were immunostained 4 days post-transduction in NCH421k cells. A significant decrease in the nMFI of CD133 was observed (Supplementary Figures S3B and C). E-NPP1-deficient GSCs are additionally characterised by increased expression of the astrocytic differentiation markers GFAP (glial fibrillary acidic protein) and S100 beta (Figures 3a and b; Supplementary Figure S3A). To further confirm the functional role of E-NPP1 in the maintenance of a stem-like phenotype in GSCs, additional progenitor/stem cell markers were studied (CD15, LHX2, MSI1 and LGR5). Strikingly, E-NPP1-deficient GSCs showed a significant decrease in the nMFI of CD15 compared with non-target control cells (Figures 3c and d) as determined by flow cytometry. Furthermore, E-NPP1-deficient GSCs showed a reduced expression of LHX2, MSI1 and LGR5 compared with non-target control cells as assessed by qRT-PCR (Figure 3e; Supplementary Figure S3E). In summary, these data suggest a wide-ranging downregulation of stem cell-associated genes upon ENPP1 knockdown. To examine this in greater detail, gene expression microarray-based transcriptome analysis was conducted on NCH421k cells transduced with shNT and ENPP1-shRNA1 4 days post-transduction and significance analysis of microarray (SAM) revealed 2104 differentially regulated genes (FDR<0.05). Using gene set enrichment analysis (GSEA), we tested an enrichment of the CD133-up signature24 and the consensus stemness ranking-up (CSR-up) signature25 in the 2104 differentially regulated genes ranked by their mean linear fold change. The CD133-up signature comprises genes (n=76) upregulated in the CD133+ GSCs compared with their autologous CD133− counterparts, whereas the CSR-up signature contains genes (n=70) upregulated in a broader array of stem cell populations reflecting a common stem cell signature. GSEA demonstrates a significant enrichment of both stem cell-associated gene sets in the most downregulated genes upon knockdown (FDR q-value<0.001; 57 genes out of 76 CD133-up associated genes; 56 genes out of 70 CSR-up signature genes; Figure 3f; Supplementary Table S1). These data allow the conclusion of an overall downregulation of stem cell-associated genes upon ENPP1 knockdown and strengthen the conclusion that E-NPP1 has an important role in the maintenance of a stem-like phenotype in GSCs.
ENPP1 knockdown induces differentiation of GSCs. (a) Western blot analysis showing a marked E-NPP1 protein knockdown in NCH421k cells and a concomitant increase in GFAP expression. Protein levels were analysed 2–4 days after transduction with shNT, ENPP1-shRNA1 (sh1) and -shRNA3 (sh3). Fold changes of protein expression relative to non-target control cells after normalisation to the α-tubulin loading control are indicated. (b) FACS analysis of GSCs (NCH421k and NCH441) transduced with shNT and ENPP1-shRNA1 (sh1) showed an increase in S100 beta upon knockdown. Quantification of FACS results was done using the nMFI (**P<0.01, n=4). (c and d) E-NPP1-deficient GSCs (NCH421k and NCH441) exhibiting reduced CD15 level as determined by flow cytometer analysis in contrast to non-target control cells. (c) Exemplary histograms of GSCs transduced with shNT and ENPP1-shRNA-3 are shown. (d) Quantification of FACS results was done using the nMFI. Mean values and S.D. are shown (*P<0.05, n=3). (e) Messenger-RNA expression analysis revealed reduced expression of MSI1 and LHX2 after ENPP1 knockdown assessed by quantitative real-time polymerase chain reaction in two GSC lines (NCH421k and NCH441). Relative expression levels were determined as described in the material and methods section. Mean values and S.D. are shown (** P<0.01, n=3). (f) Gene set enrichment analysis (GSEA) revealed an overall downregulation of stem cell-associated genes in E-NPP1-deficient GSCs. The analysis was performed with the CD133-up signature (n=76; Yan et al.24) and the CSR-up signature (n=70; Shats et al.25). The graph on the bottom represents the significantly differential expressed genes between NCH421k cells transduced with shNT and ENPP1-shRNA1 (n=2104) in a rank order based on their mean linear fold change. Genes showing an upregulation after ENPP1 knockdown are colour-coded in red and genes showing a downregulation are colour-coded in blue. The green curve corresponds to the running sum of ES that reflects the degree to which the stem cell-associated signatures (represented by the vertical lines) are overrepresented at the top or bottom of the ranked list. The NES and FDR q-value are also shown
ENPP1 knockdown reduces proliferation, induces cell death, affects cell cycle progression and increases chemosensitivity
ENPP1 is a member of the ectonucleotide pyrophosphatase/phosphodiesterase (E-NPP) family. It encodes a plasma membrane-bound enzyme cleaving pyrophosphate and phosphodiester bonds of a variety of substrates, including nucleotides (for example, ATP). Hence, E-NPP1 is implicated in a wide range of processes including nucleotide recycling, purinergic receptor signalling and ATP-mediated apoptosis.26 To further elucidate the functional consequence of ENPP1 knockdown in GSCs, we tested the 2104 differentially regulated genes for enrichment in KEGG pathways using GSEA. Lists of the top enriched KEGG pathways (FDR q-value<0.05) and of enriched genes are shown in Supplementary Table S2. Notably, knockdown of ENPP1 affects genes involved in purine and pyrimidine metabolism, which represent pathways with a pivotal role in cell proliferation by providing the building blocks for DNA and RNA synthesis. To confirm the effect of ENPP1 knockdown on purine and pyrimidine metabolism, their level was investigated by high-performance liquid chromatography (HPLC) with ultraviolet detection 2 days post-transduction. As depicted in Figure 4, knockdown of ENPP1 resulted in a global reduction of nucleosides and nucleotides. Furthermore, E-NPP1-deficient GSCs are characterised by downregulation of genes involved in DNA replication as well as cell cycle (Supplementary Table S2). As differentiation processes and cell cycle progression have been shown to be temporally coupled events,27 we further investigated the potential impact of E-NPP1 on cell growth of GSCs. Microscopic images 4 days post-transduction showed that non-target transduced GSCs efficiently formed neurospheres with regular rim and well-defined spherical shape. In contrast, knockdown of ENPP1 led to impaired neurosphere formation, with smaller spheres appearing as cell clusters with uneven rims (Figure 5a; Supplementary Figure S4). These observations were in line with a significant increase in cell death (Figure 5b; Supplementary Figure S4B) and a significant reduction in proliferation (Figure 5c) 3 days post-transduction. Moreover, cell cycle analysis upon ENPP1 knockdown revealed an accumulation of cells in G1/G0 phase (Figures 5d and e) in NCH421k cells studied 3 days post-transduction. As differentiation of GSCs was shown to increase apoptotic response to chemotherapeutic agents,15, 16 we tested whether knockdown of ENPP1 sensitised the cells to the cytotoxic substance 1,3-Bis(2-chloroethyl)-1-nitrosourea (BCNU). For this purpose, non-target control cells and E-NPP1-deficient GSCs were exposed to BCNU (50 and 75 μM) for 72 h. As depicted in Figure 5e and Supplementary Figure S4C, ENPP1 knockdown alone resulted in a significant increase in caspase-3/7 activity. Moreover, the combination of ENPP1 knockdown and BCNU treatment significantly increased apoptosis rate. Overall, these observations corroborate a role for E-NPP1 as an upstream regulator of cellular factors involved in controlling cell cycle progression and survival of GSCs.
ENPP1 knockdown decreases intracellular nucleotides and nucleosides. Determination of intracellular nucleoside and nucleotide levels in NCH421k cells transduced with shNT and ENPP1-shRNA1 (sh1) measured by HPLC and ultraviolet detection at 254 nm. Data are presented as peak area corrected for the cell number. *: not detectable
ENPP1 knockdown reduces proliferation, induces cell death, affects cell cycle progression and increases chemosensitivity. (a) Representative images showing impaired sphere forming capacity in E-NPP1-deficient NCH421k cells compared with non-target control cells. Arrowheads point to neurospheres. Insets show neurospheres at × 2.5 magnification. Scale bar=200 μM. (b) Assessment of dead cell fraction after ENPP1 knockdown in NCH421k (left bar chart) and NCH441 (right bar chart) GSCs by 7-AAD/Annexin V staining and flow cytometry study. The bar graph shows mean values and S.D. of annexin V-positive (apoptotic and/or necrotic) cells (*P<0.05; **P<0.01, n=3). (c) Assessment of proliferating cell fraction after ENPP1 knockdown in NCH421k (left bar chart) and NCH441 (right bar chart) GSCs by EdU-incorporation and flow cytometry study. Mean values and S.D. are shown (**P<0.01, n=3). (d) FACS-based cell cycle analysis of living cells revealed a G1/G0 cell cycle arrest in the E-NPP1-deficient NCH421k cells compared with non-target control cells. Nuclear DNA was stained with propidium iodide. Mean values and S.D. are shown (*P<0.05; **P<0.01, n=4). (e) Exemplary histograms of FACS measurement shown in (d). (f) Assessment of apoptosis in E-NPP1-deficient NCH421k cells upon treatment with BCNU (50 and 75 μM) by caspase-3/7 activity assay. The bar graph shows mean values and S.D. (*P<0.05, n=3). n.s.: not significant, RLU: relative light units
Expression of E2F1 and its target genes is reduced in E-NPP1-deficient GSCs
To identify potential transcription factors (TFs) responsible for the observed phenotypic effects in E-NPP1-deficient GSCs, an in silico promoter analysis based on the conducted expression profiling was performed. Genes significantly deregulated by more than 2.5-fold were considered for the analysis. Using the notion of the total binding affinity (TBA) of a promoter for a TF,28 the set of genes downregulated upon ENPP1 knockdown was found to have the highest overall binding affinity for E2F1 (FDR 3.0e-04; Supplementary Table S3), whereas no significant TF was found for the upregulated genes. To further demonstrate the role of E2F1 in the observed phenotypic effects in E-NPP1-deficient GSCs, we tested the 2104 differentially regulated genes for enrichment in experimentally validated direct E2F1 transcriptional target genes extracted from the MetaCore database (https://portal.genego.com/). Strikingly, GSEA revealed a significant enrichment of E2F1 transcriptional target genes in the most downregulated genes upon knockdown (FDR q-value<0.001; Figure 6a; Supplementary Table S1). Validation of the microarray data was achieved by qRT-PCR analysis of TK1, CDC6, PCNA and HEY1, classical E2F1 target genes facilitating G1/S phase cell cycle transition, on GSCs 4 days post-transduction using two independent shRNAs in two patient-derived GSC lines (Figure 6b, Supplementary Figure S5). To find possible TFs regulating stem cell marker expression, analysis of TBA of ‘core enrichment genes’ (defined as those genes that account for the enrichment signal, Supplementary Table S1) of the CD133-up signature as well as the CSR-up signature was conducted. The set of downregulated stem cell-associated genes was found to have a significantly high overall binding affinity for E2F1 (FDR 2.00E-04; Supplementary Table S4). The overlap of the downregulated CD133-up and CSR-up signature genes with the downregulated E2F1 target genes obtained by the gene set enrichment analyses showed that 19 out of 91 stem cell-associated genes are indeed direct E2F1 target genes (P=4.573E-12, Fisher’s Exact test; Figure 6c). In line with the proposed role of E2F1 for the observed phenotypic changes, knockdown of ENPP1 in GSCs lead to downregulation of E2F1 protein expression itself analysed 4 days post-transduction (Figure 6d). These observations suggest that E2F1 contributes to the GSC phenotype by regulating stem cell marker gene expression.
Expression of E2F1 and its target genes is reduced in E-NPP1-deficient GSCs. (a) Gene set enrichment analysis (GSEA) revealed downregulation of E2F1 target genes in E-NPP1-deficient GSCs. The analysis was performed using a set of 160 experimentally validated direct E2F1 target genes (activating interactions) from the MetaCore database (https://portal.genego.com/). The significantly differential expressed genes between NCH421k cells transduced with shNT and ENPP1-shRNA1 (n=2104) are rank ordered based on their mean linear fold change. Genes showing an upregulation after ENPP1 knockdown are colour-coded in red and genes showing a downregulation are colour-coded in blue. The green curve corresponds to the running sum of ES that reflects the degree to which the signature (represented by the vertical lines) is overrepresented at the top or bottom of the ranked list. The NES and FDR q-value are also shown. (b) qRT-PCR analysis confirmed the decreased expression of TK1, CDC6, PCNA and HEY1 after ENPP1 knockdown in NCH421k. Relative expression levels were determined as described in the Material and Methods section. Mean values and S.D. are shown (*P<0.05, **P<0.01, n=3). (c) Venn diagram depicting the overlap between downregulated CD133-up and CSR-up signature genes and downregulated E2F1 target genes upon ENPP1 knockdown. (d) Western blot analysis showing downregulation of E2F1 protein expression after ENPP1 knockdown in GSCs (NCH421k and NCH441). Fold changes of protein expression relative to non-target control cells after normalisation to the α-tubulin loading control are indicated
Discussion
One formidable challenge in improving glioblastoma therapy is its substantial cellular heterogeneity. The discovery of a rare and highly aggressive sub-population of tumour cells with stem-like features led to insights into the underlying tumour biology. Several reports implicate GSCs in therapy resistance and showed that efficacy of therapy can be improved, if stem-like cells are targeted additionally.15, 16, 29 These findings emphasise the need to improve understanding of the biological behaviours of GSCs and their underlying regulatory mechanisms. In that respect, a previous study identified kinases and phosphatases relevant for the survival of GSCs and thereby potential therapeutical targets for glioblastoma.30 Moreover, a kinome-wide RNAi screen in GSCs points to a pivotal role of phosphorylation processes as a regulatory mechanism of GSC self-renewal.15 In this study, we investigated the role of phosphatases in the maintenance of a stem-like phenotype in GSCs. Even though there is evidence for the existence of a tumourigenic CD133-negative GSC population, the stem cell-related marker CD133 has proven to be useful to isolate and further characterise tumourigenic GSCs.4, 10, 14, 24 Analysis of the extracellular CD133 epitope AC133 led to the identification of 12 phosphatases (∼7% hit rate) that significantly decreased CD133 level upon knockdown, and hence genes potentially required for the GSC phenotype. Among the candidate genes PPEF1, PTPN7, PPP4C and PPP2R5E were previously shown to be important regulators of CD133 level emphasising the validity of our screen.31 Moreover, several genes are reported to be relevant for stem cells, such as PPP4C identified in a previous RNAi screen for stem cell self-renewal factors,21 DUSP5 described as an anti-differentiation regulator in embryonic stem cells19 and PPEF1 identified as a marker for CD133+ GSCs.7 The selected candidate genes were validated in a secondary screen in three patient-derived GSC lines, but at variable level, which is most likely because of the molecular heterogeneity of tumour-derived patient samples. Among the genes involved in cell surface AC133 expression, we selected ENPP1 for further functional studies as it was shown to be overexpressed in human astrocytic brain tumours compared with control tissues and its expression strongly correlates with the glioma WHO grade.17 This membrane-bound enzyme has broad substrate specificity cleaving phosphodiester, pyrophosphate and phosphosulphate bonds of nucleotides and nucleotide derivatives.26, 32 Originally, E-NPP1 was discovered on the surface of mouse B-lymphocytes33 and is expressed in a large variety of human tissues including brain.26 Here we describe an elevated ENPP1 expression in GSCs compared with normal brain extracts and fNSCs. E-NPP1-deficient GSCs are characterised by downregulation of known stem cell markers, such as CD133, LHX2, MSI1, LGR5 and CD15,4, 8, 9, 34, 35 as well as an overall downregulation of stem cell-associated genes as seen by comparison with published stem cell gene signatures.24, 25 Moreover, GFAP and S100 beta, differentiation marker delineating the astrocytic cell lineage, are upregulated upon knockdown of ENPP1 in cultured GSCs. Hence, the change in expression of the studied markers upon ENPP1 knockdown suggests a switch towards a differentiated phenotype. Accordingly, E-NPP1-deficient GSCs showed an increased apoptotic susceptibility to the chemotherapeutic agent BCNU. A number of studies in stem cells have pointed to a crucial relationship between cell cycle progression and cell fate (self-renewal versus differentiation).36, 37, 38 Neural stem and progenitor cells are in particular characterised by a short G1 cell cycle phase and growing evidence suggests a crucial role for lengthening of G1 phase in differentiation.27 In line with this hypothesis, a number of experiments in GSCs describe co-occurrence of induced differentiation and reduced proliferation,16, 34, 39 in particular by the accumulation of cells in G1/G0 cell cycle phase.15, 40 In agreement with these findings, ENPP1 knockdown in GSCs was found to cause a reduction in the proliferation rate and cells were found to accumulate in G1/G0 cell cycle phase. To unravel the underlying cause of the observed phenotypes, an in silico promoter analysis of the deregulated genes upon ENPP1 knockdown was conducted. Investigation of the downregulated genes revealed the highest overall binding affinity for E2F1, a TF controlling G1 to S phase progression. No significant TF was found for the upregulated genes, most likely because upregulation of those genes represents an indirect effect of ENPP1 knockdown. In line with the promoter analysis, E2F1 expression and its target genes were found to be downregulated upon ENPP1 knockdown. Although a stem cell relevant role of E2F1 was so far not investigated in GSCs, E2F1 was shown to be of particular importance in the maintenance of neural stem cells41 and human pluripotent stem cells42 as well as in a transcriptional regulatory network governing the maintenance of embryonic stem cells.43 Given the central role of E2F1 in the regulation of cell cycle progression, it is reasonable to assume that the observed reduced proliferation and accumulation of cells in G1 cell cycle phase in E-NPP1-deficient GSCs is mediated by a decreased transcriptional function of E2F1.
Moreover, promoter analysis of the downregulated stem cell-associated genes in E-NPP1-deficient GSCs suggests that E2F1 has an important role in the GSC phenotype. In line with this finding, about 20% of the downregulated stem cell-associated genes are downregulated E2F1 target genes. Other TFs apart from E2F1 could not be linked to the expression of stem cell-associated genes. This might be due to more downstream effects of the knockdown. Our data suggest that E2F1 is a major mediator in maintaining GSC phenotype. More importantly, apart from promoting cell cycle progression, E-NPP1 influences expression of stem cell-associated genes by acting upstream of E2F1 transcriptional function.
E-NPP1 is – among other functions – involved in nucleotide recycling, a process particularly important in the central nervous system as de novo synthesis is mainly achieved in the liver.44 In line with this, our conducted expression profiling revealed a great deregulation of genes involved in purine and pyrimidine metabolism and nucleoside–nucleotide profiling demonstrated an altered nucleotide–nucleoside composition in E-NPP1-deficient GSCs. A balanced nucleotide pool was shown to be crucial for DNA replication.45 In addition, extracellular nucleosides and nucleotides influence proliferation, differentiation and apoptosis.46, 47 Recently, the important role of extracellular ecto-nucleotidase to balance a pool of brain nucleotides and nucleosides was brought into focus.44 Moreover, alterations in ecto-nucleotidase enzymes was linked with malignancy of solid tumours.48 In particular, alterations in activity and expression of ecto-nucleotidase were suggested to be involved in glioma progression.49 Notably, a surface-located nucleotide pyrophosphatase (most likely E-NPP1) had been identified as the major enzyme responsible for the metabolism of extracellular ATP in glioma.50 As suggested by Aerts and colleagues, highly proliferating cells require an excess of purine and pyrimidine nucleotides, hence the overexpression of ENPP1 in glioblastoma tumours is likely due to its function in nucleotide recycling.17 Our expression data strongly support this hypothesis and further links it to stem cells in glioblastoma.
We therefore hypothesise that an altered nucleotide pool in E-NPP1-deficient GSCs leads to the decreased transcriptional function of E2F1 and thereby to reduced proliferation by the accumulation of cells in G1 phase and interrelated differentiation. This imbalance in nucleotide pools might ultimately lead to apoptosis as observed previously.51
In conclusion, our data suggest that expression of ENPP1 is indispensable for the maintenance of GSCs in vitro and thus required to keep GSCs in an undifferentiated, proliferative state.
Material and Methods
Cell culture conditions and patient characteristics
The investigated GSC lines (NCH421K, NCH441 and NCH644) were established from primary glioblastoma patients undergoing surgical resection according to the research proposals approved by the Institutional Review Board at the Medical Faculty Heidelberg. Tissues were enzymatically dissociated and derived cells were cultured as floating neurospheres in serum-free DMEM/F-12 medium supplemented with 20% BIT-admixture 100 (Pelobiotech, Planegg, Germany), basic fibroblast growth factor (Biomol, Hamburg, Germany) and epidermal growth factor (Life Technology, Carlsbad, CA, USA) at 20 ng/ml each. Phenotypic and genotypic characterisations of the GSC lines were carried out in previous studies.16, 52 Patient characteristics of all samples used for RNA expression analysis are summarised in Supplementary Table S5. For passaging and plating, NCH421k and NCH441 cells were dissociated using accutase (Sigma-Aldrich, St. Louis, MO, USA) and NCH644 cells by the use of a trypsin-EDTA solution (Life Technologies, Darmstadt, Germany). Unless otherwise stated, accutase was used to dissociate GSCs before staining and flow cytometry analysis. To test the effect of BCNU (Sigma-Aldrich) a final concentration of 50 and 75 μM BCNU dissolved in DMSO, respectively, was added to the culture 1 day post-transduction for 72 h. As solvent control, DMSO was used in the same volume as 75 μM BCNU at a maximum concentration of 0.03%.
Small- and large-scale lentivirus production
The shRNA library was obtained from Sigma-Aldrich targeting 177 human phosphatases with an average of 4.1 shRNAs per gene. The lentivirus particles were produced in a 96-well format (small scale) or 6-cm petri dishes (large scale) according to the TRC (The RNAi Consortium) protocol (http://www.broadinstitute.org/rnai/public/resources/protocols). In brief, HEK293T cells were cotransfected with the plko.1 constructs and the packaging plasmids (pMD2.G, psPAX2) using Trans-IT transfection reagent (Mirus, Madison, WI, USA). Viral containing medium was collected 48 and 72 h post-transfection. For the large-scale lentivirus production viral particles were concentrated by ultra-centrifugation of the HEK293T supernatant. The titer was determined by FACS using plko.1-TurboGFP control plasmid and ranged from 2 × 105 to 5 × 105 viral particles per μl. shNT was used as negative control. ENPP1-shRNA1 ((sh1): TGAGGGACGATCTTTGAATAT) and ENPP1-shRNA3 ((sh3): CTGCGAAAGTATGCTGAAGAA)) were used for the functional studies.
CD133 antibody screen
The CD133 antibody screen was performed in non-treated 96-well plates seeding 17 000 dissociated NCH421k cells per well in a final volume of 200 μl. To minimise potential edge effects in the phenotypic assay, the outermost wells of the culture plates were filled with media only. Each plate harboured three wells with negative control (shNT), two wells with positive control (shRNA against TRRAP: GCTACGATTCTGGTGGAATAT), one well with untreated NCH421k cells (Mock) and three wells with 10 μM ATRA-treated GSCs. Neurospheres were transduced with an MOI of 5, leading to transduction efficiencies between 70 and 90%. 24 h after transduction fresh stem cell medium was added. Using a flow cytometer (FACSArray BD Biosciences, San Jose, CA, USA) CD133 levels were measured 6 days post-transduction by staining GSCs for CD133/1 using a R-Phycoerythrin (PE) conjugated antibody (clone AC133, 1.7 μg/ml; Miltenyi Biotec, Bergisch-Gladbach, Germany). A trypsin-EDTA solution (Life Technologies) was used to singularise neurospheres before the antibody staining. The MFI of live cells (using forward scatter (FSC) and side scatter (SSC)) was analysed. To account for technical variability the MFI of each sample was normalised to the mean MFI of negative (shNT) and positive controls (shTRRAP) of the corresponding plate (set to 1 and 0, respectively). A modified permutation based t-statistic was used to identify genes with a significant change in CD133 level (53 R package: siggenes). We performed a two class analysis between individual shRNA treatments (n=2–5 per gene) and negative controls of the same plate (mean MFI per well if corresponding controls were distributed over two 96-well plates). Complete permutations (n=56) were performed to estimate the null distribution. A FDR threshold of <0.05 was used to identify significantly changed genes.
Immunostaining
CD15, CD133 and S100 beta staining was performed on dissociated GSCs 3 and 4 days post-transduction, respectively, using CD15-FITC (clone VIMC6, 3.85 μg/ml), CD133/1-PE (clone AC133, 1.5 μg/ml), CD133/2-PE (clone AC141, 7 μg/ml, all Miltenyi) and S100 beta (clone SH-B1, 0.5 μg/ml, Abcam, Cambridge, UK) antibodies. Isotype control for CD15 and S100 beta was used at the same concentration, CD133 isotype control was used according to the manufacturer’s instructions. The S100 beta primary antibody was unconjugated and detected by FITC-conjugated anti-mouse secondary antibody (Dianova GmbH, Hamburg, Germany), which was used at a concentration of 1 : 200. For the immunostaining of extracellular surface markers (CD15 and CD133) the MFI of live cells (using FSC and SSC) was analysed. Before immunostaining of S100 beta fixation and permeabilisation was performed using BD Cytofix/Cytoperm kit (BD Biosciences, San Jose, CA, USA). The gate of the live cell measurement was applied and the MFI of those cells determined. Results are expressed as nMFI (nMFI=MFI (antigen-stained sample)/MFI (isotype control)). Cells were subjected to flow cytometry analysis using a FACSCanto II equipped with the FACSDiva software (BD Biosciences, San Jose, CA, USA). FlowJo software was used to overlay histograms, which were normalised to the maximum count (% of Maximum).
Nucleoside–nucleotide profiling
NCH421k cells were transduced with shNT and ENPP1-shRNA1 and nucleosides and nucleotides were extracted 2 days post-transduction following the protocol of NovoCIB (Lyon, France). In brief, cells were collected and washed with ice-cold PBS. The PBS was removed and pure ice-cold acetonitril added to the cell pellet. After extensive vortexing, the samples were incubated for 10 min on ice. The extracts were centrifuged (3000 r.p.m., 10 min, 4 °C) and the supernatant was recovered. Cold 80% acetonitrile in water (v/v) was added to the pellet and again extensively vortexed, followed by 10 min incubation at room temperature. After centrifugation (1000 r.p.m., 5 min, 4 °C) the supernatant was also transferred into the recovery tube and stored at −20 °C. Nucleoside–nucleotide profiling based on HPLC and ultraviolet detection at 254 nm was performed at NovoCIB. Data were normalised to the number of viable cells of non-target control cells as determined by trypan blue dye exclusion assay of technical replicates using Vi-CELL counter (Beckman Coulter, Fullerton, CA, USA).
Cell death
The extent of cell death was studied 3 days post-transduction and detected by flow cytometry using 7-amino-actinomycin (7-AAD) and annexin V staining. Dissociated GSCs were incubated in annexin V-binding buffer containing 10% 7-AAD and 10% annexin V-PE (BD Biosciences, Heidelberg, Germany) for 15 min at 4 °C in the dark. Immediate flow cytometry analysis was carried out using a FACSCanto II and data were analysed with FACSDiva software (BD Biosciences, San Jose, CA, USA). Annexin V-positive cells including annexin V single-positive cells (early apoptosis) and annexin V/7-AAD double-positive cells (late apoptosis and/or necrosis) were counted as dead cells.
Proliferation
Proliferation was analysed using the Click-it EdU cell proliferation assay (Life Technology) according to the manufacturer’s protocol. Briefly, 10 μM EdU was added to the culture medium 48 h post-transduction. Incorporated EdU was detected after 24 h of incubation by conjugation of Alexa Fluor 488 and subsequent flow cytometry on FACSCanto II (BD Biosciences, San Jose, CA, USA). Data were analysed using FACSDiva software (BD Biosciences, San Jose, CA, USA).
Apoptosis
For detection of apoptosis after BCNU treatment of non-target control cells and E-NPP1-deficient GSCs the activity of caspase-3/7 was analysed. To this end, GSCs were plated in technical triplicates at 8.5 × 103 per well in a final volume of 100 μl in white-walled uncoated 96-well plates (Greiner Bio-One, Kremsmünster, Austria) and transduced as described above. A set of wells was prepared with stem cell medium only for background subtraction. Following 24 h incubation, BCNU was added to the culture medium and additionally incubated for 72 h before apoptosis was assessed using luminescent caspase-3/7 activation assay according to the manufacturer’s protocol (Caspase-Glo 3/7 assay, Promega, Mannheim, Germany). In brief, an equal volume of the Caspase-Glo reagent was added to each well and the enzymatic activity of caspase-3/7 was measured using a Mithras LB 940 Microplate Reader (Berthold Technologies, Bad Wildbad, Germany).
Cell cycle
Cell cycle analysis was carried out 3 days post-transduction by propidium iodide staining as described previously.54 The cell cycle distribution was investigated on living cells by gating cells having ⩾2 N DNA content. Flow cytometry analysis was carried out using a FACSCanto II and data analysis with FACSDiva software (BD Biosciences, San Jose, CA, USA).
Western blot analysis
Cell lysates were separated in 4–12% polyacrylamide precast gels (Novex, San Diego, CA, USA) under reducing conditions and transferred onto a PVDF membrane. Total adult normal brain protein was used as control (BioChain Institute Inc., Hayward, CA, USA). Primary antibodies α-tubulin (clone DM 1A; Sigma-Aldrich), E-NPP1 (Cell signalling, Danvers, MA, USA), GFAP (clone GA5; Chemicon International, Temecula, CA, USA) and E2F1 (clone KH95; Santa Cruz Biotechnology, Santa Cruz, CA, USA) were used at a concentration of 1 : 2000, 1 : 1000, 1 : 1000 and 1 : 200, respectively. Horseradish peroxidase-conjugated secondary antibodies (Abcam) were used at a concentration of 1 : 5000 (mouse) and 1 : 10 000 (rabbit). Immunoreactive bands were detected with Pierce Fast Western Blot Kit, ECL Substrate (Thermo Scientific, Langenselbold, Germany) or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific).
RNA extraction and quantitative real-time PCR analysis
RNA from cultured cells was isolated using the RNeasy Mini kit (Qiagen, Hilden, Germany) and examined for integrity using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Total normal brain RNA pooled from five adult donors was purchased (BioChain Institute Inc.) and used as control. 1 μg of total RNA was reverse-transcribed with Superscript II (Life Technology) according to the manufacturer’s protocol. To avoid DNA contamination, RNA was pre-treated with DNaseI (Life Technology). Each complementary DNA sample was analysed in technical triplicate with the ABI PRISM 7900HT thermal cycler (Applied Biosystems, Foster City, CA, USA) using Absolute SYBR Green ROX Mix (ABgene, Epsom, UK). For relative quantification mean ratios were calculated between the gene of interest and two housekeeping genes (DCTN2 and SDHA) based on the standard curves generated by serial dilution of cDNA sample from either commercial human total RNA (Stratagene, La Jolla, CA, USA) or NCH421k RNA. For visualisation and statistical testing of the knockdown results the expression levels of the experiments were normalised to the mean expression values of individual experiments. Then, all adjusted values were normalised to the mean non-target control expression level. Oligonucleotide sequences are available in Supplementary Table S6.
Microarray analysis
NCH421k cells were transduced in three independent experiments with shNT and ENPP1-shRNA1 and RNA was extracted 4 days post-transduction. Gene expression profiling was performed using the 4 × 44K Whole Human Genome Oligo Microarray (Agilent Technologies, Wilmington, DE, USA) according to the manufacturer’s recommendations (Publication Number: G4140-90050 v.5.7, Agilent Technologies) including dye-swap on each sample to avoid dye-specific biases. Microarray slides were scanned on an Agilent Microarray Scanner G25505B (Agilent Technologies), and data were extracted with Agilent Feature Extraction software version 9.1 (Agilent Technologies). Processing of raw data including normalisation of the arrays and quality control were performed with our in-house developed ChipYard framework for microarray data analysis (http://www.dkfz.de/genetics/ChipYard/) using R statistical software and Bioconductor.55 Detailed information about the preprocessing procedure of the raw data is available upon request. SAMs53 as implemented in MeV v4.8.1 (Saeed et al.56) was applied to determine statistical significance of differential gene expression and FDR from three independent experiments. The delta value in SAM was adjusted to obtain a maximum number of significant genes (q-value<0.05) while maintaining a FDR<0.05.
Expression profiles of adherent GSCs derived from classic glioblastoma patients (G144 and G144ED (51-year-old male), G166 (74-year-old female) and GliNS2 (54-year-old male)) and of fNSCs (hf240, hf286 and hf289) were generated on GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix;23 GSE15209). Data analysis and visualisation were conducted by the use of the open access database R2 (http://r2.amc.nl). Expression data was MAS5.0 normalised before analysis.
To compare gene expression of the selected candidate phosphatases (PPEF1, ENPP1, PTPN9, PPP4C, PPP2R2A and DUSP5) in primary glioblastoma samples and adult normal brain samples, a publicly available glioma expression data set (GSE18166, Toedt et al.22) was used. Preprocessed mRNA expression data of a subset of 40 IDH1 wild-type primary glioblastoma samples were normalised to the mean expression values of four adult normal brain samples.
Gene set enrichment analysis
GSEA utilising the pre-ranked tool was performed at http://www.broadinstitute.org/gsea/57 using default settings. Significant differentially expressed genes between shNT and ENPP1-shRNA1 transduced samples were pre-ranked based on mean fold change and analysed for the enrichment of the following gene sets: CD133-up signature,24 CSR-up signature,25 direct E2F1 targets (MetaCore database, https://portal.genego.com/) and KEGG pathways (gene set c2.cp.kegg, version v3.1) as compiled in the Molecular Signatures Database.57 The gene symbols of the signature genes were updated and annotated for HUGO symbols; duplicates were removed. Genes of the signatures were matched with the normalised, processed genes on the 4 × 44K Whole Human Genome Oligo Microarray. Finally, 76 genes (CD133-up signature), 70 genes (CSR-up signature) and 160 genes (direct E2F1 targets) were retained. A gene list of the signature genes are listed in Supplementary Table S1. The GSEA method measures the enrichment of gene sets (for example, stem cell-associated signature gene sets) near the top or bottom of an ordered gene list. Quantification is achieved using a running-sum statistic and called the enrichment score (ES). The normalised ES (NES) accounts for differences in gene set size. The FDR q-value (the probability that a gene set with a given normalised enrichment score represents a false-positive finding) was used to set a significant threshold.
Computational prediction of TFs for genes deregulated by knockdown of ENPP1
To identify potential TFs important for the phenotypic effects of ENPP1 knockdown, a search for high TF affinity in the set of 377 highly downregulated genes (fold change <−2.5) was performed. To further evaluate TF with a particular influence on stem cell marker expression, analysis of TF affinity to the 57 ‘core enrichment genes’ (defined as those genes that account for the enrichment signal, Supplementary Table S1) of the CD133-up signature as well as CSR-up signature was conducted. For each of 130 non-redundant vertebrate TF matrices from the JASPAR core database,58 all genes were ranked according to their TBA28 of their promoter (defined as −1500 bp to +500 bp of the transcription start site), using the procedure described previously.59 The rank product60 of the set of 377 downregulated genes, as well as of the ‘core enrichment genes’, was taken as a score for the overall set-binding affinity of the given TF (where lower rank products indicate higher overall set-binding affinities). Finally, the 130 set-binding affinities were compared with those obtained for 10 000 random gene sets of the same size to compute FDRs and identify a list of TFs for which the set-binding affinity is significantly better than expected by chance (FDR<0.05).
Statistical analysis
Statistical analyses were performed with SigmaPlot, version 12.3 for Windows and Excel software.
Unpaired t-test was used to evaluate the significance of the differences between the relative mRNA expression values of non-target control GSCs and E-NPP1-deficient GSCs. Paired t-test was performed to determine significance of immunostaining, cell death, proliferation, cell cycle and apoptosis results comparing non-target control transduced cells and ENPP1-shRNA transduced cells. Unless otherwise specified, mean values with a S.D. of three independent experiments are presented. A P<0.05 was considered to represent significance (*) and P<0.01 was considered to be highly significant (**).
Abbreviations
- GSC:
-
glioblastoma stem-like cell
- RNAi:
-
RNA interference
- nMFI:
-
normalised median fluorescence intensity
- qRT-PCR:
-
quantitative real-time PCR
- FDR:
-
false discovery rate
- ENPP1:
-
ectonucleotide pyrophosphatase/phosphodiesterase 1
- fNSCs:
-
fetal neural stem cells
- shNT:
-
non-target shRNA
- HPLC:
-
high-performance liquid chromatography
- SAM:
-
significance analysis of microarray
- GSEA:
-
gene set enrichment analysis
- CSR:
-
consensus stemness ranking
- TBA:
-
total binding affinity
- TF:
-
transcription factor
References
Louis D, Ohgaki H, Wiestler O, Cavenee W, Burger P, Jouvet A et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 2007; 114: 97–109.
Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJB et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005; 352: 987–996.
Ignatova TN, Kukekov VG, Laywell ED, Suslov ON, Vrionis FD, Steindler DA . Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia 2002; 39: 193–206.
Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J et al. Identification of a cancer stem cell in human brain tumors. Cancer Res 2003; 63: 5821–5828.
Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res 2004; 64: 7011–7021.
Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006; 444: 756–760.
Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir I et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer 2006; 5: 67.
Son MJ, Woolard K, Nam D-H, Lee J, Fine HA . SSEA-1 Is an enrichment marker for tumor-Initiating cells in human glioblastoma. Cell Stem Cell 2009; 4: 440–452.
Toda M, Iizuka Y, Yu W, Imai T, Ikeda E, Yoshida K et al. Expression of the neural RNA-binding protein Musashi1 in human gliomas. Glia 2001; 34: 1–7.
Beier D, Hau P, Proescholdt M, Lohmeier A, Wischhusen J, Oefner PJ et al. CD133(+) and CD133(−) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res 2007; 67: 4010–4015.
Wang J, Sakariassen PØ, Tsinkalovsky O, Immervoll H, Bøe SO, Svendsen A et al. CD133 negative glioma cells form tumors in nude rats and give rise to CD133 positive cells. Int J Cancer 2008; 122: 761–768.
Pallini R, Ricci-Vitiani L, Banna GL, Signore M, Lombardi D, Todaro M et al. Cancer stem cell analysis and clinical outcome in patients with glioblastoma multiforme. Clin Cancer Res 2008; 14: 8205–8212.
Zeppernick F, Ahmadi R, Campos B, Dictus C, Helmke BM, Becker N et al. Stem cell marker CD133 affects clinical outcome in glioma patients. Clin Cancer Res 2008; 14: 123–129.
Brescia P, Ortensi B, Fornasari L, Levi D, Broggi G, Pelicci G . CD133 is essential for glioblastoma stem cell maintenance. Stem Cells 2013; 31: 857–869.
Wurdak H, Zhu S, Romero A, Lorger M, Watson J, Chiang C-y et al. An RNAi screen identifies TRRAP as a regulator of brain tumor-initiating cell differentiation. Cell Stem Cell 2010; 6: 37–47.
Campos B, Wan F, Farhadi M, Ernst A, Zeppernick F, Tagscherer KE et al. Differentiation therapy exerts antitumor effects on stem-like glioma cells. Clin Cancer Res 2010; 16: 2715–2728.
Aerts I, Martin JJ, De Deyn PP, Van Ginniken C, Van Ostade X, Kockx M et al. The expression of ecto-nucleotide pyrophosphatase/phospho- diesterase 1 (E-NPP1) is correlated with astrocytic tumor grade. Clin Neurol Neurosurg 2011; 113: 224–229.
Si-Tayeb K, Noto FK, Nagaoka M, Li J, Battle MA, Duris C et al. Highly efficient generation of human hepatocyte–like cells from induced pluripotent stem cells. Hepatology 2010; 51: 297–305.
Chen Q, Zhou Y, Zhao X, Zhang M . Effect of dual-specificity protein phosphatase 5 on pluripotency maintenance and differentiation of mouse embryonic stem cells. J Cell Biochem 2011; 112: 3185–3193.
Liu Q, Nguyen D, Dong Q, Shitaku P, Chung K, Liu O et al. Molecular properties of CD133+ glioblastoma stem cells derived from treatment-refractory recurrent brain tumors. J Neurooncol 2009; 94: 1–19.
Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ . A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev 2009; 23: 837–848.
Toedt G, Barbus S, Wolter M, Felsberg J, Tews B, Blond F et al. Molecular signatures classify astrocytic gliomas by IDH1 mutation status. Int J Cancer 2011; 128: 1095–1103.
Pollard SM, Yoshikawa K, Clarke ID, Danovi D, Stricker S, Russell R et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell 2009; 4: 568–580.
Yan X, Ma L, Yi D, Yoon JG, Diercks A, Foltz G et al. A CD133-related gene expression signature identifies an aggressive glioblastoma subtype with excessive mutations. Proc Natl Acad Sci USA 2011; 108: 1591–1596.
Shats I, Gatza ML, Chang JT, Mori S, Wang J, Rich J et al. Using a stem cell-based signature to guide therapeutic selection in cancer. Cancer Res 2011; 71: 1772–1780.
Bollen M, Gijsbers R, Ceulemans H, Stalmans W, Stefan C . Nucleotide pyrophosphatases/phosphodiesterases on the move. Crit Rev Biochem Mol Biol 2000; 35: 393–432.
Salomoni P, Calegari F . Cell cycle control of mammalian neural stem cells: putting a speed limit on G1. Trends Cell Biol 2010; 20: 233–243.
Molineris I, Grassi E, Ala U, Di Cunto F, Provero P . Evolution of promoter affinity for transcription factors in the human lineage. Mol Biol Evol 2011; 28: 2173–2183.
Piccirillo SGM, Reynolds BA, Zanetti N, Lamorte G, Binda E, Broggi G et al. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature 2006; 444: 761–765.
Goidts V, Bageritz J, Puccio L, Nakata S, Zapatka M, Barbus S et al. RNAi screening in glioma stem-like cells identifies PFKFB4 as a key molecule important for cancer cell survival. Oncogene 2012; 31: 3235–3243.
Mak AB, Blakely KM, Williams RA, Penttilä P-A, Shukalyuk AI, Osman KT et al. CD133 protein N-glycosylation processing contributes to cell surface recognition of the primitive cell marker AC133 epitope. J Biol Chem 2011; 286: 41046–41056.
Zimmermann H, Zebisch M, Sträter N . Cellular function and molecular structure of ecto-nucleotidases. Purinergic Signal 2012; 8: 437–502.
Takahashi T, Old LJ, Boyse EA . Surface alloantigens of plasma cells. J Exp Med 1970; 131: 1325–1341.
Abdouh M, Facchino S, Chatoo W, Balasingam V, Ferreira J, Bernier G . BMI1 sustains human glioblastoma multiforme stem cell renewal. J Neurosci 2009; 29: 8884–8896.
Nakata S, Campos B, Bageritz J, Lorenzo Bermejo J, Becker N, Engel F et al. LGR5 is a marker of poor prognosis in glioblastoma and is required for survival of brain cancer stem-like cells. Brain Pathol 2013; 23: 60–72.
Lange C, Calegari F . Cdks and cyclins link G1 length and differentiation of embryonic, neural and hematopoietic stem cells. Cell Cycle 2010; 9: 1893–1900.
Orford KW, Scadden DT . Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet 2008; 9: 115–128.
Singh AM, Dalton S . The cell cycle and Myc intersect with mechanisms that regulate pluripotency and reprogramming. Cell Stem Cell 2009; 5: 141–149.
Chirasani SR, Sternjak A, Wend P, Momma S, Campos B, Herrmann IM et al. Bone morphogenetic protein-7 release from endogenous neural precursor cells suppresses the tumourigenicity of stem-like glioblastoma cells. Brain 2010; 133 (Pt 7): 1961–1972.
Ying M, Wang S, Sang Y, Sun P, Lal B, Goodwin CR et al. Regulation of glioblastoma stem cells by retinoic acid: role for Notch pathway inhibition. Oncogene 2011; 30: 3454–3467.
Palm T, Hemmer K, Winter J, Fricke IB, Tarbashevich K, Sadeghi Shakib F et al. A systemic transcriptome analysis reveals the regulation of neural stem cell maintenance by an E2F1–miRNA feedback loop. Nucleic Acids Res 2013; 41: 3699–3712.
Yeo HC, Beh TT, Quek JJ, Koh G, Chan KK, Lee DY . Integrated transcriptome and binding sites analysis implicates E2F in the regulation of self-renewal in human pluripotent stem cells. PLoS One 2011; 6: e27231.
Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 2008; 133: 1106–1117.
Ipata PL . Origin, utilization, and recycling of nucleosides in the central nervous system. Adv Physiol Educ 2011; 35: 342–346.
Anglana M, Apiou F, Bensimon A, Debatisse M . Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell 2003; 114: 385–394.
Neary JT, Zimmermann H . Trophic functions of nucleotides in the central nervous system. Trends Neurosci 2009; 32: 189–198.
Ulrich H, Abbracchio M, Burnstock G . Extrinsic purinergic regulation of neural stem/progenitor cells: implications for CNS development and repair. Stem Cell Rev 2012; 8: 755–767.
Bergamin LS, Braganhol E, Zanin RF, Edelweiss MI, Battastini AM . Ectonucleotidases in tumor cells and tumor-associated immune cells: an overview. J Biomed Biotechnol 2012; 2012: 959848.
Wink MR, Lenz G, Braganhol E, Tamajusuku AS, Schwartsmann G, Sarkis JJ et al. Altered extracellular ATP, ADP and AMP catabolism in glioma cell lines. Cancer Lett 2003; 198: 211–218.
Grobben B, Anciaux K, Roymans D, Stefan C, Bollen M, Esmans EL et al. An ecto-nucleotide pyrophosphatase is one of the main enzymes involved in the extracellular metabolism of ATP in rat C6 glioma. J Neurochem 1999; 72: 826–834.
Oliver FJ, Collins MKL, López-Rivas A . dNTP pools imbalance as a signal to initiate apoptosis. Experientia 1996; 52: 995–1000.
Ernst A, Hofmann S, Ahmadi R, Becker N, Korshunov A, Engel F et al. Genomic and expression profiling of glioblastoma stem cell-like spheroid cultures identifies novel tumor-relevant genes associated with survival. Clin Cancer Res 2009; 15: 6541–6550.
Tusher VG, Tibshirani R, Chu G . Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001; 98: 5116–5121.
Nicoletti I, Migliorati G, Pagliacci MC, Grignani F, Riccardi C . A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J Immunol Methods 1991; 139: 271–279.
Gentleman R, Carey V, Bates D, Bolstad B, Dettling M, Dudoit S et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004; 5: R80.
Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 2003; 34: 374–378.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550.
Bryne JC, Valen E, Tang M-HE, Marstrand T, Winther O, da Piedade I et al. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res 2008; 36: D102–D106.
Piro RM, Molineris I, Di Cunto F, Eils R, König R . Disease-gene discovery by integration of 3D gene expression and transcription factor binding affinities. Bioinformatics 2013; 29: 468–475.
Breitling R, Armengaud P, Amtmann A, Herzyk P . Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 2004; 573: 83–92.
Acknowledgements
We thank Marc Zapatka, Herbert Zimmermann and James Watson Goding for great scientific support and helpful suggestions. We thank Ivan Molineris and Paolo Provero for providing us with the preprocessed promoter-TF affinity data and Volker Ast for providing us with the preprocessed MetaCore data. We would like to acknowledge NovoCIB (Lyon, France) for the performance of the nucleoside–nucleotide profiling. JB, VH and EP were supported by the Helmholtz International Graduate School for Cancer Research. This work was supported in part by a fund of the Bundesministerium für Bildung und Forschung, BMBF (NGFNplus, #01GS0883).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by P Salomoni
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Bageritz, J., Puccio, L., Piro, R. et al. Stem cell characteristics in glioblastoma are maintained by the ecto-nucleotidase E-NPP1. Cell Death Differ 21, 929–940 (2014). https://doi.org/10.1038/cdd.2014.12
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2014.12
Keywords
This article is cited by
-
Second messenger 2'3'-cyclic GMP-AMP (2'3'-cGAMP): the cell autonomous and non-autonomous roles in cancer progression
Acta Pharmacologica Sinica (2024)
-
Exosomal circZNF800 Derived from Glioma Stem-like Cells Regulates Glioblastoma Tumorigenicity via the PIEZO1/Akt Axis
Molecular Neurobiology (2024)
-
Identification of transcriptome alterations in the prefrontal cortex, hippocampus, amygdala and hippocampus of suicide victims
Scientific Reports (2021)
-
Enzymology of extracellular NAD metabolism
Cellular and Molecular Life Sciences (2021)
-
Thymidylate synthase maintains the de-differentiated state of triple negative breast cancers
Cell Death & Differentiation (2019)