Abstract
During apoptosis Bcl-2 proteins control permeabilization of the mitochondrial outer membrane leading to the release of cytochrome c. Essential gatekeepers for cytochrome c release are the proapoptotic multidomain proteins, Bax, and Bak. The expression of Bax is upregulated upon cellular stress by the tumor suppressor p53. Despite the high functional homology of Bax and Bak, little is known about how the bak gene is regulated. To investigate its transcriptional regulation in further detail, we have analyzed a region spanning 8200 bp upstream of the bak start codon (within exon 2) for transcription factor-binding sites, and identified three p53 consensus sites (BS1–3). Reporter gene assays in combination with site-directed mutagenesis revealed that only one putative p53-binding site (BS3) is necessary and sufficient for induction of reporter gene expression by p53. Consistently, p53 induces expression of endogenous Bak. At the mRNA level, induction of Bak expression is weaker than induction of Puma and p21. Interestingly, Bak expression can also be induced by p73 that binds however to each of the three p53-binding sites within the bak promoter region. Our data suggest that expression of Bak can be induced by both, p53 and p73 utilizing different binding sites within the bak promoter.
Similar content being viewed by others
Main
The two main apoptosis pathways are triggered by death receptors or the mitochondrial release of cytochrome c. In the mitochondrial pathway, proteins of the Bcl-2 family constitute the central regulators.1, 2 They share homology in at least one of the four identified Bcl-2 homology (BH) domains and have either anti- or proapoptotic capacity. Proapoptotic proteins are further sub-divided into multidomain proteins (i.e., Bax and Bak) that contain BH1-3 and so-called BH3-only proteins that only share homology in the BH3 domain. Both types of proapoptotic proteins interact with antiapoptotic family members such as Bcl-2, Bcl-w, Bcl-xL, and Mcl-1 – either constitutively or after activation.3, 4, 5 BH3-only proteins function as sensors for cellular stress and their activity is regulated at the transcriptional level as well as by post-translational modifications. BH3-only proteins ultimately induce activation of the multidomain proteins Bax and/or Bak that oligomerize at the mitochondrial outer membrane and induce release of cytochrome c.6 Cytosolic cytochrome c associates with Apaf-1, thereby forming the apoptosome as the activation platform for caspase-9, which then cleaves and activates downstream caspases that dismantle the cell.1, 2
Owing to the mutual but also selective interaction of pro- and antiapoptotic proteins, the mechanisms underlying Bcl-2 signaling are not fully elucidated. Currently, the ‘direct activation’ and the ‘derepression’ model are discussed. The former proposes that the BH3-only proteins Bim, Bid, and Puma are necessary to directly activate Bax or Bak by transient interaction. The derepression model suggests BH3-only proteins to inactivate antiapoptotic counterparts by binding them, thereby releasing Bax and Bak from their inactivating interaction with antiapoptotic proteins.5, 7 Interestingly, the BH3-only protein Nbk/Bik, which interacts strongly with the Bax-antagonizing proteins Bcl-2 and Bcl-xL but only weakly with Mcl-1 that predominantly interferes with Bak, does not induce apoptosis in Bax-deficient cells, arguing for a derepression mechanism.8 In addition to the specific interaction pattern of Bcl-2 proteins, their relative expression level influences the cellular response to apoptosis inducing stimuli. Expression of the BH3-only proteins Puma and Noxa is induced by p53 in response to DNA damage.9, 10 p53 also upregulates the downstream-functioning protein Bax,11 resulting in a cooperative sensitization to apoptosis. Furthermore, p53 negatively influences the expression of the antiapoptotic protein Bcl-2, leading to enhanced expression of Bcl-2 in p53-deficient cells.12, 13, 14
As bax is a p53 target gene, we wondered whether also expression of Bak, the other proapoptotic multidomain protein, is regulated by p53. We therefore analyzed the human bak1 gene locus for transcription factor-binding sites. Indeed, we identified a functional p53 consensus-binding site, and verified p53-mediated regulation of the bak1 promoter region by various means. Bak upregulation in response to DNA damage occurred later than p53-mediated induction of p21 expression and cell cycle arrest. Our data suggest a model in which cells in sustained cell cycle arrest are sensitized for apoptosis by enhanced Bak expression that shifts the rheostat of anti- and proapoptotic Bcl-2 proteins.
Results
Identification of putative p53-binding sites in the bak1 gene locus
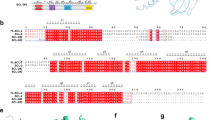
The tumor suppressor p53 suppresses Bcl-2 expression, whereas it induces several BH3-only proteins, such as Puma and Noxa, and the proapoptotic multidomain protein Bax.9, 10, 11, 12 This prompted us to analyze the possibility that p53 in addition to Bax also regulates the expression of the homologous protein Bak. We initially analyzed 8200 bp of the bak1 gene locus on chromosome 6p21.31 for potential promoter regions using the ‘Gene2Promoter’ and ‘ElDorado’ software (http://www.genomatix.de). These 8200 bp span a region that starts 5550 bp upstream of exon 1, includes exon 1, intron 1, and the first 16 codons of the coding sequence within exon 2. The software indicated two potential promoter regions, the first starting upstream of and extending into exon 1 (−3188 to −2515 bp), and the second starting within intron 1 and extending into exon 2 (−508 to 50 bp; Figure 1a). On the basis of these findings, we analyzed a total of 3350 bp (−3300 to 50 bp) including both promoter regions for transcription factor-binding sites using the ‘MatInspector’ (Genomatix, Munich, Germany) software. The software identified sites for different transcription factors, for example, SP1/2, STAT, NF-κB, and also p53 (Figure 1a). As p53 transactivates expression of several proapoptotic Bcl-2 proteins, we analyzed the three potential p53-binding sites in this region − the first one directly upstream of exon 1 and the other two following exon 1. To test whether these putative p53-binding sites were functional, we cloned the 770 bp region containing all three potential binding sites and exon 1 (−2790 to −2020) into the pGL3-basic vector generating pGL3-p53BS, and performed luciferase reporter gene assays.
Representation of the bak1 gene locus and reporter gene assays. (a) Schematic representation of the analyzed region of the bak1 gene locus on 6p21.31. The promoter regions predicted by the Gene2Promoter and ElDorado software, positions of exon 1, intron 1, and exon 2 and positions of the three putative p53-binding sites are depicted. Also shown is the sequence of the cloned bak promoter region. The putative p53-binding sites are printed in bold and highlighted by gray boxes, the position of exon 1 is highlighted in gray, and potential binding sites for SP1 and STAT are underlined. (b) Luciferase reporter gene assay using the reporter vector containing the illustrated region (−2790 to −2020 bp) of the bak1 gene locus. HeLa and HCT116/p53−/− cells were co-transfected with pGL3-Basic-p53BS together with different amounts of a p53 expression vector. Tumor suppressor p53 induces luciferase expression in a concentration-dependent manner in HeLa and HCT116/p53−/− cells. Values are means±S.D. from six independent experiments
p53 induces target gene expression from bak promoter constructs
We co-transfected HeLa cells with different amounts of a p53 expression vector, together with the reporter plasmid pGL3-p53BS, containing all three putative p53-binding sites. Clearly, co-transfection of p53 concentration dependently induced luciferase expression from the bak-promoter at 48 h after transfection (Figure 1b). These data were confirmed by similar experiments in HCT116/p53−/− cells, in which exogenous p53 also specifically induced luciferase activity from the cloned region of the bak1 gene locus (Figure 1b).
To investigate the specific role of each of the three putative p53-binding sites, we generated four additional luciferase reporter vectors containing either mutations in one or each of the three putative p53-binding sites. The p53 consensus site consists of two half sites of the sequence 5′-PuPuPuC(A/T)(A/T)GPyPyPy-3′ separated by 0–13 bp.15 We introduced mutations in each of these sites separately by exchanging the essential cytosine and guanine residues for adenine and thymine (Supplementary Table 1). Additionally, we created a reporter vector in which all three p53-binding sites were mutated simultaneously. These four reporter gene plasmids, pGL3-Basic-mutBS1, −mutBS2, −mutBS3, and −mutBS1-3 were then co-transfected together with the p53 expression construct into HeLa cells. Unexpectedly, mutation of BS1 or BS2 resulted in negligible changes of luciferase activity compared with the non-mutated construct, whereas mutation of BS3 caused a significant reduction of luciferase activity by about 60% (Figure 2a). Mutation of BS3 had a similar effect on HCT116/p53−/− cells. Again, mutation of BS1 or BS2 did not significantly influence reporter gene expression, whereas luciferase activity induced from pGL3-Basic-mutBS3 was reduced by about 60% (Figure 2b). The simultaneous mutation of all three putative p53-binding sites did not further reduce luciferase activity compared with the single mutation of BS3 in either of the cell line. In agreement with the previous results, induction of luciferase expression was higher in HeLa than in HCT116/p53−/− cells. These data demonstrate that p53 induces target gene expression from the bak1 promoter and, moreover, that exclusively BS3 is necessary for p53-driven gene expression, whereas BS1 and BS2 are dispensable.
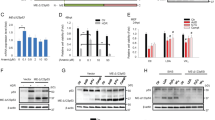
Identification of BS3 as the functional p53-binding site. The HeLa (a) and HCT116/p53−/− (b) cells were transfected with the indicated reporter gene constructs together with a vector for the expression of p53. Luciferase reporter gene induction by p53 is not affected by mutation of BS1 or BS2, whereas mutation of BS3 reduces luciferase activity significantly. (c) Electrophoretic mobility shift assays using the p53 consensus sequence (p53con), BS1, BS2, BS3, or mutated BS3 (mutBS3), and extracts from etoposide-treated HCT116/wt and HCT116/p53−/− cells. Addition of a p53 antibody produces a supershift (indicated by asterisk) of p53con and BS3 in samples from wildtype but not from p53-deficient cells. No shift of BS1, BS2, or mutBS3 is detected. (d) Anti-p53 ChIP assay from HCT116/wt cells incubated in the absence or presence of 50 μM etoposide for 72 h. The graph shows enrichment of BS3 but not BS1 and BS2 over input DNA. Values are averages of two independent experiments. (e) HCT116 cells were transduced with 10 or 50 MOI of adenoviruses for the expression of p53 or p14ARF. After 3 days, cells were collected and total RNA analyzed by qRT-PCR. Expression of p53 in HCT116/p53−/− and p14ARF in HCT116/wt cells induces Bak mRNA expression, whereas p14ARF does not trigger upregulation of Bak mRNA in HCT116/p53−/− cells. Values represent means±S.D. from at least three independent experiments
p53 directly binds to BS3
As exclusively mutation of BS3 reduced reporter gene activity, we next investigated p53 binding in electrophoretic mobility shift assays (EMSAs) using ds-oligonucleotides containing the sequence of each putative binding site. We also included the mutated BS3 and the p53 consensus-binding site in these assays. The radiolabeled probes were incubated with nuclear extracts from HCT116/wt cells that had been treated for 48 h with etoposide. In samples containing the p53 consensus site, a protein/DNA complex was readily detected and addition of an anti-p53 antibody produced a supershift, proving the identity of the p53/DNA complex (Figure 2c). Although each of the binding site oligonucleotides was unexpectedly shifted in EMSAs exclusively the BS3 oligonucleotide was supershifted by anti-p53. Mutation of BS3 abolished protein binding in the presence or absence of the p53 antibody. Hence, p53 directly binds to BS3, and mutation of the essential cytosine and guanine residues in the BS3 sequence disrupts p53 binding. We further performed EMSAs, with extracts from etoposide-treated HCT116/p53−/− cells, and also detected protein/DNA complexes of each of the binding sites; however, addition of anti-p53-Ab did not produce a supershift. These results confirm direct binding of p53 to BS3, but also show that BS1-3 are bound by additional proteins. Finally, we performed chromatin immunoprecipitation (ChIP) to analyze binding of p53 to BS1, BS2, and BS3 within the bak promoter in a cellular context. In anti-p53 ChIPs from etoposide-treated HCT116/wt cells we found enrichment of BS3, but not BS1 or BS2 (Figure 2d), thus confirming binding of endogenous p53 specifically to BS3 within the bak promoter.
p53 upregulates expression of endogenous Bak mRNA
The previous results demonstrate that p53 induces target gene expression from bak promoter constructs by binding to BS3. Consequently, we investigated endogenous bak gene expression and transduced HCT116/p53−/− cells with different MOIs of an adenovirus encoding for p53. The mRNA levels of Bak were elevated (about sixfold after 3 days) in Ad-p53-transduced cells (Figure 2e). We also aimed to achieve p53 upregulation in a more physiological manner, and therefore transduced HCT116/wt and HCT116/p53−/− cells with an adenovirus encoding for p14ARF. p14ARF inhibits MDM2-mediated p53 degradation and induces elevated levels of p53. Enhancing the level of endogenous p53 clearly resulted in elevated levels (>10-fold) of Bak mRNA in HCT116/wt cells (Figure 2e). The specificity of these results was underlined by transduction of HCT116/p53−/− cells, in which p14ARF did not induce Bak mRNA expression (Figure 2e).
We then investigated whether activation of endogenous p53 in response to DNA damage drives Bak expression. We incubated HCT116/wt cells and, as a control, isogenic HCT116/p53−/− cells with etoposide, and then analyzed Bak mRNA expression by quantitative real-time PCR (qRT-PCR). The level of Bak mRNA was upregulated over time in etoposide-treated HCT116/wt cells (Figure 3a), that is, Bak expression was increased by 1.75-fold after 24 h and increased up to 4.5-fold at 72 h. In HCT116/p53−/− cells, treated with etoposide, induction of Bak mRNA expression was less pronounced (1.5-fold at 48 h and 2.7-fold at 72 h; Figure 3a).
Time course of p53-dependent gene induction in response to DNA damage. The HCT116/wt and HCT116/p53−/− cells were treated with 50 μM etoposide for 24, 48, and 72 h and expression of Bak (a), p21 (b), Puma (c), and Bax (d) mRNA was analyzed by qRT-PCR. The mRNA expression of Bak, Bax, Puma, and p21 rises over time in response to treatment with etoposide in HCT116/wt and to a lesser extent in HCT116/p53−/− cells. Values are means±S.D. from three independent experiments
The EMSA results indicated binding of proteins different from p53 to BS1, BS2, and BS3, which are reflected by Bak mRNA induction in p53-negative cells. To test whether upregulation in the absence of p53 is Bak specific, we investigated induction of known p53 target genes, such as the cyclin-dependent kinase inhibitor p21, Bax, and the BH3-only protein Puma.9, 11, 16, 17 In line with the literature, qRT-PCR analysis showed a time-dependent induction of all target genes in response to etoposide treatment of HCT116/wt cells. Induction was strongest for p21 reaching a 55-fold expression level at 72 h (Figure 3b). Induction of Puma was below that of p21 (25-fold at 72 h, Figure 3c) and that for Bax was weakest (sevenfold at 72 h, Figure 3d). Comparable to the results obtained for Bak, all these established p53 target genes were also induced in p53-deficient HCT116/p53−/− cells, albeit to a lesser extent. We therefore conclude that bak can be considered a bona fide p53 target gene.
We also tested whether the p53-mediated induction of target genes correlates with the strength of DNA damage. To this end, we treated HCT116/wt and HCT116/p53−/− cells for 48 h with different doses of etoposide, resulting in a concentration-dependent bak induction in HCT116/wt and to a lesser extent in HCT116/p53−/− cells (Figure 4a). For comparison, we also analyzed mRNA induction of Bax, p21, and Puma. In HCT116/wt cells, induction of Bax mRNA was comparable to that of Bak (Figure 4b), and induction of Puma mRNA expression was higher than that of Bax and Bak (up to 4.5-fold, Figure 4c). Notably, p21 was induced about 16-fold regardless of the etoposide concentration (Figure 4d). The multidomain protein Bax and Bak mRNA expressions were significantly reduced in HCT116/p53−/− cells compared with HCT116/wt cells (Figures 4a–b). Expression of p21 and Puma mRNA after etoposide treatment was slightly elevated in HCT116/p53−/− cells (Figures 4c–d).
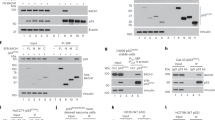
Analysis of mRNA and protein expression of p53 target genes in response to etoposide. HCT116/wt and HCT116/p53−/− cells were treated with 10 μM, 50 μM, and 100 μM etoposide for 48 h and expression of Bak (a), Bax (b), Puma (c), and p21 (d) mRNA was analyzed by qRT-PCR. Expression of Bak and Puma mRNA increases with concentration of etoposide, whereas induction of Bax and p21 mRNA expression is independent of the concentration of etoposide. (e) The HCT116/wt and HCT116/p53−/− cells were incubated with 50 μM etoposide for 24 h, 48 h and 72 h, and expression of p53, Bak, Bax, and p21 was analyzed by western blotting. (f) The HCT116/wt and HCT116/p53−/− cells were incubated with the indicated concentrations etoposide for 48 h and expression of p53, Bak, Bax, and p21 protein was analyzed likewise. In HCT116/wt and to a lesser extent also in HCT116/p53−/− cells, the level of Bak expression rises in a time- (e) and concentration- (f) dependent manner. Values are means±S.D. from three independent experiments
p53-dependent upregulation of Bak protein level in response to DNA damage
We demonstrated p53-specific regulation of bak gene expression, and hence analyzed Bak protein expression in cells that had been treated with etoposide from 24 h to 72 h. Confirming our qRT-PCR results, Bak protein was strongly upregulated by etoposide in HCT116/wt cells, and only weakly in HCT116/p53−/− cells (Figure 4e). Expression of p21 protein was already highly induced at 24 h of etoposide treatment and then slightly declined over time. Western blot analysis also confirmed p53-dependent induction of Bax (Figure 4e). Interestingly, compared with wildtype cells, HCT116/p53−/− cells had reduced basal levels of Bak expression. This Bak expression in HCT116/wt cells is presumably caused by a basal amount of p53, whereas the minor Bak expression in HCT116/p53−/− cells is presumably due to other transcription factors. In summary, expression of p21 and Bax was already induced by low doses of etoposide, whereas Bak expression increased in a dose-dependent manner (Figure 4f).
p73 transactivates Bak
Having established p53-dependent upregulation of the proapoptotic protein Bak in response to DNA damage, we were left with the interesting finding that Bak expression, albeit to a lower extent, is also upregulated in the absence of p53. Bak induction in the absence of p53 could be because of redundancy of the p53 family proteins regarding their consensus binding sites. Because p73 has been shown to induce expression of Puma in response to γ-irradiation,18 Bak expression might also be affected by p73. To test this, we co-transfected HCT116/p53−/− cells with expression plasmids for different p73 isoforms together with the bak promoter construct. All p73 isoforms (α, β, γ, δ) induced luciferase expression to various degrees (data not shown). The isoform p73δ showed the highest induction and was used in subsequent experiments. We next co-transfected the p73 expression vector together with the unmutated and the different mutated bak promoter constructs. The p73-induced luciferase expression from the bak promoter region (Figure 5a) was reduced by mutation of each of the putative p53-binding sites to 70–80%, whereas mutation of all three binding sites had an additive effect and reduced reporter gene expression by about 60%. Hence, bak gene expression is induced by p73, and this induction is regulated by all three binding sites rather than exclusively by BS3 as is the case for p53. Also in EMSAs, addition of a p73 antibody produced a supershift of all unmutated oligonucleotides used, again showing that p73 binds to BS1, BS2, and BS3 (Figure 5b).
The p73-mediated induction of bak-promoter driven reporter gene expression, DNA binding, and repression by Δ133p53, DNp73, and p73 knockdown. (a) The HCT116/p53−/− cells were co-transfected with the indicated reporter gene plasmids and a p73α expression vector. After 48 h cells were collected and luciferase activity was assessed. Values are expressed as fold induction normalized to the activity induced from pGL3-Basic alone. Reporter gene induction was reduced by the single mutation of each of the putative p53-binding sites, whereas mutation of all three binding sites had an additive effect. (b) The HCT116/p53−/− cells were treated with 50 μM etoposide for 48 h and EMSAs were performed on nuclear extracts using 32P-labeled ds-oligonucleotides for the p53 consensus sequence (p53con), BS1, BS2, BS3, and mutated BS3 (mutBS3). Addition of a p73 antibody produces a supershift (*) of p53con, BS1, BS2, and BS3, thus indicating binding of p73α to these oligonucleotides. (c) The HeLa and HCT116/p53−/− cells were co-transfected with p73 and Δ133p53 or DNp73 together with the bak promoter reporter construct. After 48 h, cells were collected and luciferase activity detected luminometrically. Induction of luciferase activity by p73 was reduced to basal level in cells co-transfected with the DNp73 expression plasmid, whereas no reduction was seen in Δ133p53 co-expressing cells. (d) The HeLa and HCT116/p53−/− cells were co-transfected with p53 and Δ133p53 or DNp73 together with the bak promoter reporter construct. Induction of luciferase activity by p53 was measured at 48 h after transfection and reduced in both Δ133p53 and DNp73 co-expressing cells. (e) The HCT116/p53−/− cells were transduced with 50 MOI of Ad-p73α and at 72 h, the samples were analyzed for Bak mRNA expression by qRT-PCR. The Bak mRNA expression was upregulated by 2.5-fold in Ad-p73 transduced cells compared with control cells. (f) The HCT116/wt and HCT116/p53−/− cells were transfected with a vector for the expression of p73-shRNA and incubated in the absence or presence of 50 μM etoposide for 72 h. The qRT-PCR revealed inhibition of etoposide-induced Bak mRNA expression by the p73-specific shRNA as compared with the control in both wild-type and p53-deficient HCT116 cells
Both p53 and p73 can also be expressed as N-terminal truncated isoforms that lack the transactivation domain, and function dominant negative on their respective transactivation-competent TA isoforms.19 We investigated the effect of ΔN isoforms on p73-mediated reporter gene induction by co-expression of Δ133p53 and DNp73. As expected, p73-induced reporter expression from the bak promoter was reduced by DNp73 in HeLa and HCT116/p53−/− cells to basal level (Figure 5c). Interestingly, p73-mediated reporter gene induction was not reduced by co-expression of Δ133p53. In complementary experiments, we investigated the influence of DNp73 and Δ133p53 on p53-induced reporter gene expression. In HeLa cells, reporter gene induction by p53 was reduced by 70% when DNp73 was co-expressed, and by 30% in case of Δ133p53 co-expression (Figure 5d, left panel). Likewise, in HCT116/p53−/− cells expression of DNp73 diminished p53-mediated luciferase expression from 12.5- to 2.2-fold, and Δ133p53 again showed less reduction of p53-induced luciferase activity (12.5-fold versus 7.6-fold) (Figure 5d, right panel).
We finally tested whether p73 induces expression of endogenous Bak and transduced HCT116/p53−/− cells with 50 MOI of an adenovirus encoding for p73α. At 72 h after transduction, qRT-PCR showed a 2.5-fold increase of Bak mRNA in p73-expressing HCT116/p53−/− cells as compared with control cells (Figure 5e). Furthermore, p73-mediated upregulation of Bak mRNA in response to etoposide treatment was analyzed in HCT116/wt cells, transfected with an p73-shRNA expression vector, and control cells. Expression of p73 was upregulated by fourfold in etoposide-treated control cells and only by 1.5-fold in shp73-transfected cells (data not shown). In line, the eightfold induction of Bak mRNA in etoposide-treated cells was reduced to about fourfold in p73-shRNA-expressing cells (Figure 5f), again verifying p73-mediated upregulation of endogenous Bak. To further investigate whether p73 regulates Bak mRNA expression, we also incubated HCT116/p53−/− cells that had been transfected with p73-shRNA in the absence or presence of etoposide for 3 days. Upregulation of Bak mRNA was reduced from 2.3-fold in etoposide-treated cells to 1.7-fold in p73-shRNA-expressing cells (Figure 5f). These results confirm that p73 can modulate Bak mRNA expression independently of p53.
To finally investigate the relevance of Bak expression for DNA damage-induced apoptosis, we compared sub-G1 populations in HCT116/wt, HCT116/Bax−/−, HCT116/Bakknockdown, and HCT116/Bax−/−/Bakknockdown cells treated with etoposide for up to 72 h (Figure 6a). As expected, Bax knockout and Bak knockdown reduced the number of hypodiploid, and hence apoptotic cells at each time point (Figure 6b). Interestingly, after 72 h of treatment, combined knockout of Bax and knockdown of Bak had an additive effect that was not evident after 24 h and 48 h. At this late time point, apoptosis was reduced by about 50% in HCT116/Bax−/−/Bakknockdown, and only about 25% in either HCT116/Bax−/− or HCT116/Bakknockdown compared with HCT116/wt cells.
Apoptosis induction in Bak- or Bax-deficient and proficient HCT116 cells. (a) Western blot analyses show the status of Bax and Bak expression in the individual cell lines; actin served as loading control. (b) The indicated Bax- or Bak-proficient and deficient HCT116 cell lines were incubated with 50 μM etoposide for up to 72 h and DNA content of cell nuclei was analyzed by flow cytometry. Nuclei with less than 2 N of DNA content (sub-G1) were quantified as apoptotic. Knockout of Bax protects HCT116 cells more efficiently from apoptosis than knockdown of Bak at 24 h and 48 h. At later time points, combined Bak knockdown and Bax knockout have an additive protective effect
Discussion
We identified the proapoptotic protein Bak as a transcriptional target of p53, and we have furthermore, for the first time, characterized the bak1 promoter region with respect to p53-binding sites. We found that p53-mediated reporter gene upregulation exclusively depends on the p53-BS3 spanning from bp −2085 to −2066. Binding of p53 to this region was verified by EMSAs and ChIP assays. Induction of Bak mRNA and protein by p53 was confirmed by qRT-PCR, and western blot analyses in HCT116/wt cells and the isogenic p53 knockout cell line. Hence, like other Bcl-2 family proteins, for example, Puma, Noxa, Bax, and Bcl-2, also Bak is transcriptionally regulated by p53. The location of the p53 response element downstream of exon 1 within the first intron is reportedly a common feature of some p53 target genes, such as GADD45α, KILLER/DR5, and MDM2.15, 20, 21, 22
Our qRT-PCR data show that Puma induction by p53 is stronger than that of either Bak or Bax, but still does not reach the level of p21. This reflects that cell cycle regulatory genes contain more robust p53REs than do proapoptotic genes.23 The strength of p53-mediated induction of the proapoptotic genes puma, bax, and bak1 correlates with the distance of their respective p53REs from the transcriptional start site, as is the case for many p53 target genes.24 The puma p53RE is located in the promoter region (−145/−126), those for bax (354/373), and bak1 (596/617, corresponding to BS3) are located in the first intron. Although other mechanisms might regulate p53 target gene expression, the relative induction of these proteins reflects the cellular response that is established by p53. At first strong induction of p21 induces cell cycle arrest. Then under sustained cell cycle arrest and as long as Puma expression is insufficient to induce apoptosis, the proapoptotic proteins Bax and Bak accumulate. Finally, cells cross the borderline to apoptosis because of the overall shift in the Bcl-2 rheostat, resulting in cell death rather than cell cycle arrest.
In addition to p53, p73 can drive gene expression from the bak promoter. We found that p73 induces reporter gene expression from the bak promoter, and that transgenic expression of p73 and also etoposide treatment induce Bak expression in p53-negative HCT116 cells. Interestingly, all three investigated binding sites equally contributed to p73-mediated luciferase expression, whereas p53-mediated reporter gene induction exclusively depended on BS3. An overlapping response element affinity of p53 and p73 has already been shown by Osada et al.25 However, in addition to binding to p53 consensus sites, p73 has also its own specific consensus sequence.25 The presence of additional specific p73 consensus sites in the bak promoter region is presumable, as mutation of all three putative p53-binding sites did not abrogate p73-driven reporter gene activity.
We also found that ΔN isoforms differentially influence p53- and p73-induced reporter gene expression. Although DNp73 suppresses reporter gene induction by both p53 and p73, the ΔN133p53 isoform specifically reduces gene induction by p53 but not by p73. However, the effect of ΔN133p53 and DNp73 on transactivation by p53 and p73 not only depends on binding specificity and affinity, but also on the relative expression level of transcription factors. Therefore, transactivation of the bak promoter by endogenous p53- and p73 isoforms might differ to some degree from results of reporter gene assays.
The expression of several Bcl-2 family proteins has been shown to be induced by transcription factors that do not belong to the p53 family. Expression of Puma for instance can also be induced by FoxO3 and SP1.26, 27 Interestingly, Chirakkal et al.28 identified a SP1-binding site within the bak1 promoter that was also indicated in our MatInspector analysis (Figure 1). This SP1-binding site mediates upregulation of Bak in response to butyrate as well as basal Bak expression. The presence of this SP1-binding site in our reporter gene constructs might therefore explain that mutation of p53-binding sites does not completely abrogate gene induction. Most recently, NF-κB was shown to induce expression of Puma, Noxa, Bad, and Bim, thereby contributing to apoptosis induction by TNF-α.29 As our software analysis also indicated several NF-κB-binding sites in the bak promoter, it is likely that apart from p53 other factors influence Bak expression under certain conditions. For example, in embryogenesis Wilm's tumor suppressor protein 1 (WT1) has been implicated in regulation of Bak expression.30 In summary, all these findings reflect that the regulation of Bcl-2 protein expression by numerous transcription factors is still poorly investigated. A comprehensive model for the complex mechanisms that underlie control of apoptosis by Bcl-2 proteins would need to incorporate both, transcriptional and post-transcriptional regulation.
Materials and Methods
Cell lines, reagents, and antibodies
HCT116/wt, HCT116/p53−/−, HCT116/bax−/− (kindly provided by B Vogelstein),31 and HCT116 cells stably expressing Bak siRNA (HCT116/bakknockdown and HCT116/bax−/−/bakknockdown)32 were maintained in McCoy's 5A medium, supplemented with 10% heat-inactivated fetal calf serum, 10 mM glutamine and antibiotics (100 U/ml penicillin, 0.1 mg/ml streptomycin) (PAA Laboratories, Linz, Austria). HeLa cells were cultured in RPMI-1640 (PAA Laboratories) with identical supplements. All chemicals were purchased from Sigma (Munich, Germany). The following antibodies were used: monoclonal mouse anti-Bax (6A7, Trevigen, Gaithersburg, MD, USA), polyclonal rabbit anti-Bak (Upstate, Temecula, CA, USA), monoclonal mouse anti-p53 (Ab-6, Calbiochem/EMD Biosciences, Darmstadt, Germany), monococlonal mouse anti-p73 (ER-13, Sigma), monoclonal mouse anti-p21 (Becton Dickinson, Heidelberg, Germany), and monoclonal mouse anti-β-actin (Sigma). Secondary antibodies to mouse and rabbit IgG were from Promega (Mannheim, Germany).
Cloning and mutagenesis
The bak1 promoter region was amplified by PCR from clone RPCIP704J10291Q (RZPD, Berlin, Germany) using the primer pair BS-for/BS-rev (Supplementary Table 1) that contained the XhoI and HindIII restriction sites. After restriction with XhoI and HindIII, the PCR product was ligated into the pGL3-Basic vector (Promega) generating pGL3-Basic-p53BS. Vectors containing mutations in the putative p53 consensus-binding sites 1 and 3 were generated by PCR amplification using the primer pair mutBS1-for/BS-rev and BS-for/mutBS3-rev (Supplementary Table 1), respectively. Subsequent ligation of the PCR products into pGL3-Basic resulted in pGL3-Basic-mutBS1 and pGL3-Basic-mutBS3. Binding site 2 was mutated using the Quick Change Mutagenesis kit (Stratagene, La Jolla, CA, USA). The vector pGL3-Basic-p53BS served as template and the two complementary primers mutBS2-for and mutBS2-rev (Supplementary Table 1) introduced the desired mutations, resulting in pGL3-Basic-mutBS2. To generate the vector pGL3-Basic-mutp53BS-containing mutations in all three binding sites simultaneously, the template pGL3-Basic-mutBS2 was PCR amplified using the primers mutBS1-for and mutBS3-rev. All vector sequences were confirmed by DNA sequencing.
Western blot analysis
Cells were collected by scraping, then were washed in ice-cold PBS and lysed in buffer containing 1% Nonidet P-40, 20 mM HEPES (pH 7.9), 2 mM PMSF, 350 mM NaCl, 1 mM MgCl2, 0.5 mM EDTA, 0.1 mM EGTA, and 0.5 mM DTT supplemented with complete protease inhibitor cocktail (Roche, Mannheim, Germany). Protein concentrations were determined using the BCA assay kit (Thermo Fischer Scientific, Bonn, Germany) and 15 μg of protein per lane were loaded onto standard SDS-PAGE gels and separated at 200 V. The proteins were transferred onto a polyvinylidene difluoride membrane (Amersham Bioscience GmbH, Freiburg, Germany) by tank blotting. The membranes were blocked in PBS containing 4% BSA and 0.05% Tween-20 for 1 h, followed by an overnight incubation with the primary antibody diluted in blocking buffer at 4°C. After washing the membranes thrice in blocking buffer, the appropriate secondary antibodies (1 : 5000) were applied for 1 h. Finally, membranes were washed for an additional 2 h in PBS and 0.05% Tween-20, before proteins were visualized using ECL reagents (Amersham Biosciences).
Transfection and viral transduction
Cells were seeded for 24 h before transfection in six-well plates or culture dishes at a density of 7.5 × 104 cells/cm2. Transfection was performed using the Fugene 6 (Roche) or jetPEI (PEQLAB, Erlangen, Germany) reagent according to the manufacturer's protocol. Transfected cells were grown for various time periods before collecting. Preparation of p14ARF, p53, and p73 adenoviruses and transduction of cells were performed as described.33, 34, 35
Luciferase reporter gene assays
Cells were transfected with the respective pGL3-Basic reporter plasmid together with expression vectors for p53, Δ133p53, DNp73, p73δ, or empty vector.36, 37, 38 At 48 h after transfection, cells were collected by scraping and lysed in LLB (25 mM glycylglycine (pH 7.8), 15 mM MgSO4, 4 mM EGTA (pH 8.0), 1 mM DTT, and 1% Triton X-100) for 5 min on ice. After centrifugation (5 min, 10 000 × g), 10 μl of the supernatants were assayed in 100 μl luciferase assay buffer (15 mM potassium phosphate (pH 7.8), 25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, 1 mM DTT, and 2 mM ATP) using a microplate luminometer.39 Light emission was measured after injection of 100 μl of luciferin (0.3 mg/ml).
Electrophoretic mobility shift assays
The p53-consensus oligonucleotide was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA) and the oligonucleotides representing BS1, BS2, BS3, and mutBS3 (Supplementary Table 2) were from Sigma. Hybridized oligonucleotides were radiolabelled by Klenow-reaction in the presence of [γ]-32P-ATP. Nuclear extracts (10 μg) from etoposide (50 μM, 48 h)-treated HCT116/wt or HCT116/p53−/− cells were incubated with radiolabeled oligonucleotides for 30 min at room temperature. For supershift assays, 1 μg of the respective antibody was added to the samples followed by an additional incubation for 30 min on ice. Samples were separated on an 8% of 0.5 × TBE polyacrylamide gel at a voltage of 160 V, were dried, and autoradiagraphs were taken by exposure to an X-ray film for 3–5 h.
Quantitative real-time PCR
Cells were seeded at a density of 1 × 105 cells/cm2. Twenty four hours later, the culture medium was changed for fresh medium with or without etoposide (50 μM). After the indicated periods of time, cells were collected by scraping, and total RNA was isolated using the RNeasy mini kit from Qiagen (Hilden, Germany) according to the manufacturer's protocol. Complementary DNA was reverse transcribed from 1 μg total RNA using a two-step protocol: first, RNA was incubated with 50 μM random hexamer primers (Fermentas GmbH, Leon-Rot, Germany) and 40 U RNase-Inhibitor (RiboLock RNase Inhibitor, Fermentas) at 65°C for 10 min. Then, 200 U of reverse transcriptase (Revert Aid H Minus M-MuLV Reverse, Fermentas), 10 mM DTT, 400 μM dNTPs, and water were added to make a final volume of 25 μl, and samples were incubated at 42°C for 50 min followed by final heat inactivation at 72°C for 10 min. The qPCR was performed in an ABI7300 instrument (Applied Biosystems, Darmstadt, Germany) using 1 × Maxima SYBR Green qPCR Mastermix (Fermentas), 300 nM sense/anti-sense primers, and 80 ng cDNA. The qPCR program started with an initial denaturation step at 95°C for 10 min followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. Melting curve analysis was performed by heating the samples twice to 95°C for 15 s and 60°C for 1 min. Negative control reactions contained water instead of cDNA and were included in each run to ensure absence of contamination. Primers used for analysis of p21 (Hs_CDKN1A_1_SG), puma (Hs_BBC3_1_SG), and bak (Hs_BAK1_1_SG) expression were purchased from Qiagen and primers for bax expression (forward: 5′-CACGGCAGAGAATGCCTATGA-3′, reverse: 5′-CCCAATTGATGCCACTCTCAA-3′) were kindly provided by Dr. K Lauber (University Hospital Tübingen). Results were normalized to GAPDH and analyzed by the ΔΔCt method to give fold induction as compared with untreated control samples.
Chromatin immunoprecipitation assay
Cells were seeded at a density of 1 × 105 cells/cm2. Twenty four hours later, the culture medium was changed for fresh medium with or without 50 μM etoposide. After 3 days, cells were collected by scraping, crosslinked, and ChIPs were performed from 5 × 107 cells using an anti-p53 antibody (Ab-6) as described.40 Quantitative real-time PCR was performed on a LightCycler480 instrument using the SYBR Green I Master kit (Roche) according to the manufacturer's protocol. Primers specific for amyloid precursor protein (APP), BS1, BS2, and BS3 are indicated in Supplementary Table 3. Values were normalized to APP and fold enrichment over input chromatin was calculated using the 2exp{−ΔΔCt} method.
Abbreviations
- BS:
-
binding site
- CDK:
-
cyclin-dependent kinase
- EMSA:
-
electrophoretic mobility shift assay
- qRT-PCR:
-
quantitative real-time PCR
References
Danial NN, Korsmeyer SJ . Cell death: critical control points. Cell 2004; 116: 205–219.
Schwerk C, Schulze-Osthoff K . Regulation of apoptosis by alternative pre-mRNA splicing. Mol Cell 2005; 19: 1–13.
Galonek HL, Hardwick JM . Upgrading the BCL-2 network. Nat Cell Biol 2006; 8: 1317–1319.
Kim H, Rafiuddin-Shah M, Tu HC, Jeffers JR, Zambetti GP, Hsieh JJ et al. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat Cell Biol 2006; 8: 1348–1358.
Willis SN, Chen L, Dewson G, Wei A, Naik E, Fletcher JI et al. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev 2005; 19: 1294–1305.
Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ et al. Proapoptotic Bax and Bak: a requisite gateway to mitochondrial dysfunction and death. Science 2001; 292: 727–730.
Willis SN, Fletcher JI, Kaufmann T, van Delft MF, Chen L, Czabotar PE et al. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science 2007; 315: 856–859.
Gillissen B, Essmann F, Hemmati PG, Richter A, Richter A, Oztop I et al. Mcl-1 determines the Bax dependency of Nbk/Bik-induced apoptosis. J Cell Biol 2007; 179: 701–715.
Nakano K, Vousden KH . PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell 2001; 7: 683–694.
Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T et al. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 2000; 288: 1053–1058.
Miyashita T, Reed JC . Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 1995; 80: 293–299.
Miyashita T, Krajewski S, Krajewska M, Wang HG, Lin HK, Liebermann DA et al. Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene 1994; 9: 1799–1805.
Bourgarel-Rey V, Savry A, Hua G, Carre M, Bressin C, Chacon C et al. Transcriptional down-regulation of Bcl-2 by vinorelbine: identification of a novel binding site of p53 on Bcl-2 promoter. Biochem Pharmacol 2009; 78: 1148–1156.
Sohn D, Essmann F, Schulze-Osthoff K, Janicke RU . p21 blocks irradiation-induced apoptosis downstream of mitochondria by inhibition of cyclin-dependent kinase-mediated caspase-9 activation. Cancer Res 2006; 66: 11254–11262.
el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B . Definition of a consensus binding site for p53. Nat Genet 1992; 1: 45–49.
el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM et al. WAF1, a potential mediator of p53 tumor suppression. Cell 1993; 75: 817–825.
Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B . PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell 2001; 7: 673–682.
Melino G, Bernassola F, Ranalli M, Yee K, Zong WX, Corazzari M et al. p73 Induces apoptosis via PUMA transactivation and Bax mitochondrial translocation. J Biol Chem 2004; 279: 8076–8083.
Harms KL, Chen X . The functional domains in p53 family proteins exhibit both common and distinct properties. Cell Death Differ 2006; 13: 890–897.
Kastan MB, Zhan Q, el-Deiry WS, Carrier F, Jacks T, Walsh WV et al. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell 1992; 71: 587–597.
Zauberman A, Flusberg D, Haupt Y, Barak Y, Oren M . A functional p53-responsive intronic promoter is contained within the human mdm2 gene. Nucleic Acids Res 1995; 23: 2584–2592.
Takimoto R, El-Deiry WS . Wild-type p53 transactivates the KILLER/DR5 gene through an intronic sequence-specific DNA-binding site. Oncogene 2000; 19: 1735–1743.
Weinberg RL, Veprintsev DB, Bycroft M, Fersht AR . Comparative binding of p53 to its promoter and DNA recognition elements. J Mol Biol 2005; 348: 589–596.
Riley T, Sontag E, Chen P, Levine A . Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 2008; 9: 402–412.
Osada M, Park HL, Nagakawa Y, Begum S, Yamashita K, Wu G et al. A novel response element confers p63- and p73-specific activation of the WNT4 promoter. Biochem Biophys Res Commun 2006; 339: 1120–1128.
You H, Pellegrini M, Tsuchihara K, Yamamoto K, Hacker G, Erlacher M et al. FOXO3a-dependent regulation of Puma in response to cytokine/growth factor withdrawal. J Exp Med 2006; 203: 1657–1663.
Ming L, Sakaida T, Yue W, Jha A, Zhang L, Yu J . Sp1 and p73 activate PUMA following serum starvation. Carcinogenesis 2008; 29: 1878–1884.
Chirakkal H, Leech SH, Brookes KE, Prais AL, Waby JS, Corfe BM . Upregulation of BAK by butyrate in the colon is associated with increased Sp3 binding. Oncogene 2006; 25: 7192–7200.
Wang P, Qiu W, Dudgeon C, Liu H, Huang C, Zambetti GP et al. PUMA is directly activated by NF-kappaB and contributes to TNF-alpha-induced apoptosis. Cell Death Differ 2009; 16: 1192–1202.
Morrison DJ, English MA, Licht JD . WT1 induces apoptosis through transcriptional regulation of the proapoptotic Bcl-2 family member Bak. Cancer Res 2005; 65: 8174–8182.
Zhang L, Yu J, Park BH, Kinzler KW, Vogelstein B . Role of BAX in the apoptotic response to anticancer agents. Science 2000; 290: 989–992.
Hemmati PG, Guner D, Gillissen B, Wendt J, von Haefen C, Chinnadurai G et al. Bak functionally complements for loss of Bax during p14ARF-induced mitochondrial apoptosis in human cancer cells. Oncogene 2006; 25: 6582–6594.
Hemmati PG, Gillissen B, von Haefen C, Wendt J, Starck L, Guner D et al. Adenovirus-mediated overexpression of p14(ARF) induces p53 and Bax-independent apoptosis. Oncogene 2002; 21: 3149–3161.
Zender L, Kuhnel F, Kock R, Manns M, Kubicka S . VP22-mediated intercellular transport of p53 in hepatoma cells in vitro and in vivo. Cancer Gene Ther 2002; 9: 489–496.
Gillissen B, Essmann F, Graupner V, Starck L, Radetzki S, Dorken B et al. Induction of cell death by the BH3-only Bcl-2 homolog Nbk/Bik is mediated by an entirely Bax-dependent mitochondrial pathway. Embo J 2003; 22: 3580–3590.
Graupner V, Schulze-Osthoff K, Essmann F, Janicke RU . Functional characterization of p53beta and p53gamma, two isoforms of the tumor suppressor p53. Cell Cycle 2009; 8: 1238–1248.
Grob TJ, Novak U, Maisse C, Barcaroli D, Luthi AU, Pirnia F et al. Human delta Np73 regulates a dominant negative feedback loop for TAp73 and p53. Cell Death Differ 2001; 8: 1213–1223.
Essmann F, Pohlmann S, Gillissen B, Daniel PT, Schulze-Osthoff K, Jänicke RU . Irradiation-induced translocation of p53 to mitochondria in the absence of apoptosis. J Biol Chem 2005; 280: 37169–37177.
Totzke G, Essmann F, Pohlmann S, Lindenblatt C, Jänicke RU, Schulze-Osthoff K . A novel member of the IkappaB family, human IkappaB-zeta, inhibits transactivation of p65 and its DNA binding. J Biol Chem 2006; 281: 12645–12654.
Rothfuss O, Fischer H, Hasegawa T, Maisel M, Leitner P, Miesel F et al. Parkin protects mitochondrial genome integrity and supports mitochondrial DNA repair. Hum Mol Genet 2009; 18: 3832–3850.
Acknowledgements
We thank Bert Vogelstein, Martina Müller-Schilling, Stefan Kubicka and Kirsten Lauber for providing valuable materials and helpful discussion. We also thank Mohamed Hassan for support in EMSA experiments. This work was supported by the Deutsche Forschungsgemeinschaft (SFB773, SFB685, GRK1302) and the BMBF (AID-Net).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by H-U Simon
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Graupner, V., Alexander, E., Overkamp, T. et al. Differential regulation of the proapoptotic multidomain protein Bak by p53 and p73 at the promoter level. Cell Death Differ 18, 1130–1139 (2011). https://doi.org/10.1038/cdd.2010.179
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2010.179
Keywords
This article is cited by
-
Ethyl acetate extract of Elephantopus mollis Kunth induces apoptosis in human gastric cancer cells
BMC Complementary Medicine and Therapies (2021)
-
Mithramycin selectively attenuates DNA-damage-induced neuronal cell death
Cell Death & Disease (2020)
-
Eukaryotic initiation factor 5B (eIF5B) provides a critical cell survival switch to glioblastoma cells via regulation of apoptosis
Cell Death & Disease (2019)
-
Apoptosis resistance, mitotic catastrophe, and loss of ploidy control in Burkitt lymphoma
Journal of Molecular Medicine (2015)
-
Terfenadine induces anti-proliferative and apoptotic activities in human hormone-refractory prostate cancer through histamine receptor-independent Mcl-1 cleavage and Bak up-regulation
Naunyn-Schmiedeberg's Archives of Pharmacology (2014)