Abstract
Background:
Timely diagnosis and classification of colorectal cancer (CRC) are hindered by unsatisfactory clinical assays. Our aim was to construct a blood-based biomarker series using a single assay, suitable for CRC detection, prognostication and staging.
Methods:
Serum metabolomic profiles of adenoma (N=31), various stages of CRC (N=320) and healthy matched controls (N=254) were analysed by gas chromatography-mass spectrometry (GC-MS). A diagnostic model for CRC was derived by orthogonal partial least squares-discriminant analysis (OPLS-DA) on a training set, and then validated on an independent data set. Metabolomic models suitable for identifying adenoma, poor prognosis stage II CRC and discriminating various stages were generated.
Results:
A diagnostic signature for CRC with remarkable multivariate performance (R2Y=0.46, Q2Y=0.39) was constructed, and then validated (sensitivity 85%; specificity 86%). Area under the receiver-operating characteristic curve was 0.91 (95% CI, 0.87–0.96). Adenomas were also detectable (R2Y=0.35, Q2Y=0.26, internal AUROC=0.81, 95% CI, 0.70–0.92). Also of particular interest, we identified models that stratified stage II by prognosis, and classified cases by stage.
Conclusions:
Using a single assay system, a suite of CRC biomarkers based on circulating metabolites enables early detection, prognostication and preliminary staging information. External population-based studies are required to evaluate the repeatability of our findings and to assess the clinical benefits of these biomarkers.
Similar content being viewed by others
Main
Colorectal cancer (CRC) is the third highest cause of cancer deaths in the world. Early detection and diagnosis makes cure possible. Therefore, substantial efforts have been made to devise and implement screening strategies. Currently, colonoscopy represents the gold standard screening test. However, colonoscopy is invasive, inconvenient and expensive. To enhance the effectiveness of screening strategies, stool-based tests for occult blood have been used to triage low-risk individuals for colonoscopy. However, CRC screening remains below target (Smith et al, 2015). Therefore, a blood-based screening test used in this manner could have an important clinical role.
Treatment for CRC depends on accurate staging. More advanced locoregional disease requires the addition of systemic therapy to surgery alone, and disseminated disease is typically managed by systemic therapy alone. In recent years, numerous efforts have been made to identify patients with apparent early-stage disease who might benefit from systemic therapy. For example, a number of groups have devised methods to identify occult lymph node metastases with the intent of identifying a subgroup of stage II patients who might benefit from adjuvant therapy (André et al, 2009; Croner et al, 2014). Others have taken the approach of identifying stage II patients with an adverse prognosis using various molecular profiling techniques, including proteomics and transcriptomics (Salazar et al, 2010; Kennedy et al, 2011; Roth et al, 2012).
Metabolomics is capable of characterising individuals by disease state. Moreover, in cancer the metabolome is a close molecular representation of tumour phenotype (Bathe and Farshidfar, 2014), thus it is possible to identify clinically relevant metabolomic subgroups. In recent years, several groups have attempted to characterise the metabolomic changes associated with CRC (Bertini et al, 2012; Farshidfar et al, 2012; Ma et al, 2012; Mal et al, 2012; Nishiumi et al, 2012). However, all of these studies lack a large validation cohort, and there have been few attempts at exploring potential clinical applications. The primary aim of this study was to identify and validate a metabolomic signature for CRC using blood samples from a large patient cohort. Our secondary aim was to explore the potential usefulness of metabolomic profiling for screening, prognostication and staging. Our approach involved the identification of important discriminatory metabolites with targeted analysis of these metabolites in independent validation sets.
Materials and methods
Sample collection
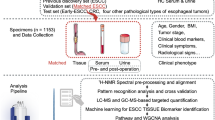
This study was approved by the Conjoint Health Research Ethics Board at the University of Calgary (Ethics ID E21805) and conforms to the Declaration of Helsinki (2008). Serum samples and clinical information were collected in a prospective cohort of consented CRC patients, treated at the Foothills Medical Center (Calgary, AB, Canada) between 2006 and 2013. Patients with extrahepatic metastases, any acute inflammatory state, sepsis and any hereditary adenomatosis syndrome (including family history with a first-degree relative with CRC) were excluded. Colorectal adenoma samples and control samples were collected prospectively by the Forzani and MacPhail Colon Cancer Screening Centre at the University of Calgary. Disease-free controls consisted of healthy individuals who had a normal screening colonoscopy. Controls were matched for age and sex with cancer and adenoma patients; ages were within 2 years, except in patients older than 75 years, where matches were within 5 years of age. Individuals were all fasting for at least 8 h before blood collection. Sera were collected in gold top Vacutainer tubes (BD Biosciences, Mississauga, ON, Canada). Samples were processed within 6 h of collection and were stored at −20 °C until the day of analysis (Hollander and Wolfe, 1999; Saeed et al, 2003).
Metabolomic analysis
Sera were divided into training sets and validation sets, and then analysed by gas chromatography-time of flight-mass spectrometry. Samples were arranged in batches that included all comparator groups, randomly distributed. That is, each batch had approximately equal representation of all stages of CRC and also contained matched disease-free controls; there was a random distribution based on sex and date of collection.
Gas chromatography-mass spectrometry
Gas chromatography-mass spectrometry methods have been previously described (Farshidfar et al, 2012). For metabolite extraction, we used a modified Bligh and Dyer extraction and purification method (Bligh and Dyer, 1959). Gas chromatography-mass spectrometry was performed using an autosampler equipped Agilent 7890A chromatograph (Agilent Technologies Canada Inc., Mississauga, ON, Canada) coupled with a Waters GCT Premier Orthogonal Acceleration/Time-of-Flight Mass Spectrometer (Waters Corp., Milford, MA, USA). An MS range of 50–800 m/z was used for scanning. Acquired spectra were processed using Metabolite-Detector software (ver. 2.1N; Technische Universität Carolo-Wilhelmina zu Braunschweig, Braunschweig, Germany). For metabolite identification, an in-house library based on the GOLM metabolome database (Hummel et al, 2007) and NIST 2011 (Stein, 1995) library, and MassBank mass spectral database (Horai et al, 2010) were used. Named metabolites are Metabolomics Standards Initiative (MSI) level 2 (putatively annotated compounds); the remainder of the compounds would be classified as MSI level 4 (unknown compounds). Retention time, retention indices, and the ions are reported for each compound. Identified peak intensities were normalised for each sample using median fold-change normalisation (Dieterle et al, 2006). Missing values were imputed with the smallest value in the data set.
Data analysis
Student’s t-test was used for statistical comparison of pairs of groups, and Fisher’s exact test was used to compare proportions. All univariate tests were two-sided, and a P-value <0.05 was considered a priori to be statistically significant. Where applicable, the significance threshold was adjusted for multiple testing, using the Holm–Bonferroni correction method. In the univariate analysis of stage-dependent variations, we took a nonparametric approach (Hollander and Wolfe, 1999) (Kruskal–Wallis test) with a Bonferroni correction in Multi-Experiment Viewer, version 4.9 (The TM4 Software Development Team, http://mev.tm4.org/) (Saeed et al, 2003). The Spearman’s rank correlation metric was used for hierarchical clustering, using complete linkage as the linkage method for generating heatmaps. To correct for analytical batch variation in GC-MS, we used the ComBat approach, originally designed for batch correction in microarray experiments and available through the ‘sva’ package (Leek et al, 2012) in Bioconductor (Gentleman et al, 2004) in R environment (ver. 3.0.2) (R Core Team, 2014). Calculated values were used as preprocessed data.
Preprocessed GC-MS data were log-transformed, centered and unit-variance scaled before being imported to SIMCA multivariate analytical software (version 14.0.0; Umetrics AB, Malmö, Sweden). Compounds with skewedness over 2.5 were further checked for analytical quality, including confirmation of correct compound identification and examination for outliers. A preliminary principal component analysis (PCA) was generated for each comparison with up to three components per PCA. Thus, intrinsic clustering and distinct patterns arising from specific metabolites’ distributions, as well as potential outlier samples, could be detected by PCA. Potentially significant subsets of metabolites were selected by using two-sample t-test assuming unequal variances (Welch’s t-test). Metabolites with a P-value <0.30 were deemed informative (Weljie et al, 2007) and were used for subsequent orthogonal partial least-squares discriminate analyses (O-PLS-DA) or O2-PLS-DA. Compounds with variable importance on projection (VIP) of less than a defined threshold were further filtered. For each analysis, the VIP threshold was set so that R2Y and Q2Y are maximised, and their difference is minimised. Based on our prior work, this variable selection approach can be used reliably for multivariate statistical comparisons, such as O-PLS-DA Further, this variable selection method has been successfully used to generate the most informative and parsimonious classification models (Weljie et al, 2007; Bathe et al, 2011; Farshidfar et al, 2012).
In assessment and comparison of O-PLS-DA models, R2Y and Q2Y scores were used to assess the variance coverage by predictive component, and predictability of the model in a seven times cross-validation, respectively (Farshidfar et al, 2012). A difference of >0.2 between R2Y and Q2Y scores was re-evaluated in each case. Fitted models were checked for a potential effect of confounders as described in the Results section. To inspect the validity and potential overfit in the PLS-based models, a 999 times permutation test was used for the outcomes of interest (Egdington, 1987). Results are presented as Q2 intercept. A Q2 intercept of zero or below demonstrates the stability and non-randomness of the model, and therefore strongly supports the validity of the model (Triba et al, 2015; UmetricsAB, 2015).
To evaluate the predictive performance of the constructed signatures in external validations, the area under the receiver-operating characteristic curves (AUROC) was calculated by GraphPad Prism (version 6.01 for Windows; GraphPad Software, La Jolla, CA USA; http://www.graphpad.com).
Pathway analysis
We used QIAGEN’s Ingenuity Pathway Analysis (IPA; QIAGEN Redwood City, http://www.qiagen.com/ingenuity) for pathway analyses of metabolites studied and identified. HMDB identifiers (Wishart et al, 2007) were used along with KEGG identifiers (Kanehisa and Goto, 2000). Metabolites identified to be of significance in the O-PLS-DA analyses were selected and a data set containing their identifiers were uploaded to the IPA. Meticulous effort was made to make sure of exact assignment from IPA’s Knowledge Base of endogenous chemicals. We then projected these metabolites onto the IPA global metabolite networks. The connectivity networks of eligible metabolites were created using the metabolomic and core analyses in IPA (using experimentally observed connections).
Results
The characteristics of the study cohort are summarised in Table 1. Samples were randomly assigned to training set and validation set. In patients with stage IVa disease, 50 (35%) received chemotherapy within 3 months before sampling, 32 patients (20%) with non-metastatic disease received chemotherapy before sampling. None of the patients had chemotherapy within 4 weeks of sampling. In all of the analyses described below, chemotherapy did not have a measurable effect on metabolomic profile. While this may be because any effects of chemotherapy had diminished in the 4-week washout period, the lack of any systematic changes in the metabolome may also be because of the diversity of chemotherapeutic drugs used.
Identification of metabolites associated with CRC
The training set consisted of 222 CRC cases (including each stage of disease) and 156 controls. Principal component analysis showed no specific clustering in relation to clinical factors; no batch-dependent effect was seen. However, there was some non-random separation between CRC and control samples (explained variance, R2X=0.25) (Figure 1A). In a supervised exploration, 41 out of 212 metabolites passed the filtering procedure before the final refinement of the metabolomic model (Supplementary Table 1). The final metabolomic model diagnostic of CRC clearly separated cases of CRC from disease-free controls: R2Y was 0.46 and Q2Y was 0.39 (cross-validation analysis of variance (CV-ANOVA) P-value <0.00001) (Figures 1B and C). The permutation test Q2 intercept was −0.12, which demonstrates that the model is stable and non-random.
The metabolomic profile of CRC patients as determined by GC-MS is distinct from disease-free controls. (A) Principal component analysis scores scatter plot of CRC and matched controls. (B) Supervised (O2PLS-DA) analysis scores scatter plot of CRC and matched controls. (C) Coefficient column plot for OPLS-DA of CRC vs matched control, illustrating changes in individual compounds. (D) Receiver-operating characteristic curve curve for validation of metabolomic classification of CRC and control, in an independent sample set (NM, not matched (unidentified)).
Metabolomic profiles are known to vary with sex (Griffin and Nicholls, 2006; Slupsky et al, 2007). Similarly, we identified sex-specific patterns (Figure 2). In males, the metabolomic model could be reduced to 48 metabolites: R2Y was 0.49 and Q2Y was 0.38 (CV-ANOVA P-value <0.00001, permutation Q2-intercept=−0.17). In females, the model could be reduced to only 17 metabolites: R2Y was 0.51 and Q2Y was 0.43 (CV-ANOVA P-value <0.00001, permutation Q2 intercept=−0.17).
The general metabolomic signature diagnostic for CRC was then subjected to validation in an independent training set consisting of 98 CRCs (with the representation from all stages) and 67 matched controls. The diagnostic signature had a promising sensitivity of 85% and a specificity of 86%. Overall, the precision of the model (positive predictive value) was over 89%. The AUROC was 0.91 (95% CI, 0.87–0.96) (Figure 1D).
Using the metabolites perturbed in CRC to populate a model, we performed a pathway analysis using IPA. The metabolic perturbations were related to a number of diseases in addition to CRC, including other digestive system cancers and hepatocellular carcinoma, as well as other epithelial neoplasias. Metabolic functions include uptake of L-alanine, D-glucose transport, threonine degradation, glycine biosynthesis, tyrosine biosynthesis and phenylalanine aerobic degradation. Highly correlated functions were growth of organism, proliferation of cells, metabolism of carbohydrate, synthesis of carbohydrate and proliferation of tumour cells. Anticorrelated functions were cell death in tumour cell lines, metabolism of proteins, necrosis, apoptosis of tumour cell lines, peroxidation of lipid, necrosis of epithelial tissue and binding of cells. The network generated on the perturbed pathways showed increased involvement of the NF-κB, PI3K, AKT-ERK1/2, MAPK and insulin-related pathways (Supplementary Figure 1). This pattern is a very similar to what we have reported previously (Farshidfar et al, 2012).
Detection of very early-stage disease
Adenoma is the preneoplastic state in the majority of sporadic colorectal adenocarcinomas. We sought to characterise the metabolomic state in the sera of average risk patients with the very early disease. For 30 out of 31 adenoma cases, only one adenoma ⩾6 mm was found on endoscopy, and one case had two adenomas. The patients were all in the age range of 50–70 years old, and sex distribution was representative of the population distribution (Table 1).
Sera from adenoma and control cases were analysed using GC-MS. After exclusion of compounds that were not consistently detected, 147 compounds were selected for further examination. Seventy-eight (53%) of the compounds could be identified using the GOLM and NIST reference libraries. All 147 compounds were subjected to multivariate analysis. In PCA, there was no clustering related to age, sex or diagnosis (Figure 3A). After filtration, 38 metabolites were incorporated into a supervised multivariate analysis (OPLS-DA). The refined fitted model included 14 compounds (R2Y=0.35; Q2Y=0.26; CV-ANOVA P-value=0.0002, permutation Q2 intercept=−0.20) (Figure 3B and Supplementary Table 2). The model was able to discriminate between the control and disease states, although there was some overlap. In estimating the performance of the model in cross-validation, the AUROC was 0.81 (95% CI, 0.70–0.92) (Figure 3C).
Metabolomic profile of colorectal adenoma, as determined by GC-MS (A–C). (A) Principal component analysis comparison of the GC-MS spectra of colorectal adenomas and disease-free controls. (B) Supervised (OPLS-DA) analysis scores scatter plot of adenomas and controls, from GC-MS spectra. (C) Receiver-operating characteristic curve curve of the GC-MS-derived biomarker for adenoma, from internal cross-validation.
Finally, we performed a targeted analysis of metabolites that were found to be progressively more perturbed as tumour burden increased, speculating that these same metabolites are also minimally altered with very early disease. Using this approach, we were not able to improve on the classifier model derived from the non-targeted analysis.
Mapping stage-related changes in the metabolome
We have previously identified that some metabolomic features are stage-related and organ-specific (Farshidfar et al, 2012). However, there is little understanding of how the circulating serum metabolome changes as CRC advances, as it invades successive layers of the bowel wall and spreads to regional lymph nodes. Although it was possible to separate stages I, II and III disease (data not shown), the seven metabolites that changed with stage did not change in a single direction with successive stages. This may be due to the fact that stage III is defined only by positive lymph node status, with heterogeneous T-stages. For this reason, the effects of primary tumour extent and lymph node status were considered separately.
First, we wanted to identify metabolites that become progressively more perturbed as the disease burden increases. T1 and T2 clustered together and were separable from T3 and T4 tumours (Figures 4A and B). The metabolomic model that separated T-stages consisted of 40 metabolites (Supplementary Table 3 and Figure 4C). In addition to identifying metabolites that change with increasing T-stage (as a categorical variable), we identified metabolites that changed progressively with increasing primary tumour dimension (analysed as a continuous variable), because T-stage may not be the best reflection of disease burden related to the primary tumour. We identified 23 metabolites that were altered in proportion to tumour size (Supplementary Table 4). These metabolites would be targeted for analysis of very early disease (adenoma), where minute changes in these same metabolites may be present. Finally, N-stage could be categorised as a metabolomic model based on 17 compounds (Figure 4D). The compounds that are included in this model are listed in Supplementary Table 5.
Metabolomic changes related to disease stage. (A) Scores scatter plot of supervised (OPLS-DA) analysis illustrating that the metabolomic profile of locoregional CRC is dependent on its T-staging status. (B) Box and whisker plot of OPLS-DA scores for each of four different T statuses. Points shown are out of the range of 2.5–97.5%. (C) Heatmap representing relative concentrations for each of the 45 compounds composing the OPLS-DA model for differentiation of T status. (D) Supervised OPLS-DA scores scatter plot representing the alterations in the metabolomic profile of lymph node-positive vs lymph node-negative CRC.
Prognostication of stage II disease
It is well known that a subgroup of patients with stage II disease has a higher risk of recurrence. This has been related to the molecular features of the tumour (Kennedy et al, 2011; Marisa et al, 2013). We wanted to explore whether the metabolomic profile could also be used to identify patients who are at high risk of recurrence. Pre-treatment of sera from 50 patients with stage II disease were analysed, based on a minimum follow-up of 36 months for patients who remained disease-free. Median follow-up was 60.3 months (range, 7.3–101.3 months). There were 12 recurrences within 36 months of resection (24% recurrence rate). The clinical features of stage II patients are summarised in Supplementary Table 6.
The metabolomic profiles of patients who recurred were markedly different from the profile of patients who did not recur (Figure 5A). The model consisted of 31 metabolites (Supplementary Table 7), which separated the group by one predictive component (R2Y=0.60; Q2Y=0.41; CV-ANOVA P<0.0001, permutation Q2 intercept=−0.24). Age, sex, degree of differentiation, tumour dimension and T-stage each had some measurable effect on the metabolome when considered in a univariate manner. However, when analysed in a multivariate model (O2PLS), sex, degree of differentiation, tumour dimension and T-stage did not have a significant influence on the model. Age was the only factor that independently had a minor effect on the separation of recurrence vs no recurrence stage II patients (R2VY=0.21).
Evaluation of the capability of the metabolomic profile to separate stage II patients by prognosis. (A) Orthogonal partial least squares-discriminant analysis (OPLS-DA) scores scatter plot demonstrating differences in the metabolomic signature in good prognosis and bad prognosis stage II patients. (B) Analysis of stage II patients to determine whether their profile is more stage I-like or stage III-like, using the OPLS-DA predictive scores derived from the model distinguishing these two stages. Red triangles represent individuals who had a recurrence.
Others have shown that some patients deemed stage II may be understaged. One reason for this may be an inadequate number of lymph nodes sampled (Sarli et al, 2005; Gleisner et al, 2013). Therefore, we examined whether any of the stage II patients had a metabolomic profile that resembled the profile of a patient with node-positive disease (Figure 5B). Although there were individuals with stage II disease who had profiles that resembled stage III disease, these were not the individuals who had recurrent disease. Therefore, we do not believe that this contributed to our observations.
Discussion
Recent studies have reported on the molecular features of CRC at the genetic, epigenetic, transcriptional and protein levels (The Cancer Genome Atlas Network, 2012). An integrated analysis has provided an important means to characterise subgroups of CRC, although no data are available to integrate associated metabolomic features, which might be considered a reflection of the phenotype. We have studied a large group of patients with various stages of CRC to identify and validate the metabolomic features that generally characterise CRC, and we have explored the utility of harnessing the metabolome to find a biomarker profile for various applications in domains currently lacking a satisfactory diagnostic approach.
Using GC-MS, we have identified an accurate and repeatable metabolomic biomarker diagnostic for CRC. The effect of each individual metabolite was not great. However, the metabolomic biomarker for CRC consists of multiple corelated metabolites perturbed as a result of pathophysiologic changes. In essence, a metabolomic biomarker is a ‘meta-biomarker’. External validation demonstrated that this pattern of change is characteristic of the disease state, and it is reproducible.
Other groups have made similar efforts in characterising the metabolomic features of CRC with varying results. Qiu et al (2009) determined metabolomic profiles in a small group of patients with CRC and healthy controls using GC-MS and liquid chromatography-mass spectrometry. In a follow-up study on a larger patient cohort, 249 metabolites were analysed using the two analytical platforms. Alterations in metabolites related to TCA cycle, urea cycle, glutamine metabolism and gut flora metabolism were reported (Tan et al, 2013). Nishiumi et al (2012) reported on a cohort of 60 Japanese individuals with CRC compared with healthy controls; sera were analysed by GC-MS. When the metabolomes from the two studies using GC-MS were compared with the present study, only one metabolite, hydroxybutyrate, was found to be consistently increased. Leichtle et al (2012) evaluated amino acids using MS/MS in 59 German patients with CRC and disease-free controls. Eleven amino acids were decreased in CRC, which differs from the amino-acid profile in our CRC patients, although we also found methionine and valine to be decreased in CRC. In summary, the metabolomic signatures generated by various investigators differ.
The variation in metabolomic signature reported by various investigators is related to a number of factors. Variations are partly related to population-based diversity (i.e., differences in genetics, environmental differences including diet, as well as differences in disease stage and molecular subtype). In addition, there are technical variations, including the analytic platforms and the actual instrumentation used. Even when the same analytical platform is used, investigators may optimise the settings differently under a standardised protocol. Therefore, it is imperative that downstream biomarker studies be accompanied by well-controlled, standardised methods to ensure repeatable results. As quantitative and targeted assays are developed, technical causes of any variability of signatures will be minimised. Finally, to address issues related to the effects of a test population on the biomarker, it will be important to perform external validation using diverse patient cohorts when metabolomic biomarkers are developed (Zhu et al, 2014).
From a diagnostic biomarker perspective, perhaps the most advanced effort was reported by Tan et al (2013). As in our study, this team reported a discovery group and a validation cohort. The top 10 metabolites identified in their study were used to generate a model that yielded a sensitivity of 83.7% and a specificity of 91.7%. However, their biomarker profile consisted of metabolites identified on two separate analytical platforms. Our best diagnostic model was derived from a single analytical modality, and it compared favorably.
Perhaps, the most exciting aspect of this work is the demonstrated capability to detect very early colorectal neoplasia in serum samples. Classifiers distinguishing solitary adenoma from disease-free controls derived from GC-MS had an AUROC of 0.81. Patients with adenoma and disease-free controls were collected uniformly, from average risk individuals undergoing screening colonoscopy. The identification of metabolomic changes associated with the size of tumour and depth of invasion enabled a targeted analysis of these same metabolites in patients with adenoma. However, this did not improve the performance of the biomarker for adenoma. While external validation is needed to fully appreciate the performance of the metabolomic blood test for adenoma (i.e., sensitivity and specificity), our findings are intriguing. As a comparison, fecal occult blood test detects 7–11% of adenomas, and fecal immunoassay test (FIT) has a sensitivity of 22–61% (Heitman et al, 2010). However, more recent studies have shown lower performance for detection of advanced neoplasm and larger adenomas by FIT (20–67%) and even lower accuracy for FOBT assays (Whitlock et al, 2008; Cubiella et al, 2014). The FIT AUROC for advanced neoplasms is about 0.7 in an average risk population (Cubiella et al, 2014). We are therefore quite encouraged by our findings. Moreover, because blood tests are more convenient than fecal tests, it is conceivable that patients will be more agreeable to serial, repeated tests, which may further enhance the detection rate.
Gas chromatography-mass spectrometry is a potentially a very attractive analytical platform for development of biomarkers destined for the clinic, for it is accurate and sufficiently sensitive (Dunn et al, 2011). Importantly, it is easy to implement and takes up a small footprint the size of a desktop, making it applicable in a clinical laboratory. Our GC-MS-based biomarker for CRC is therefore poised for continued development. The weakness of GC-MS as used here is that it is only semiquantitative. To enhance its performance (and the fidelity of biomarkers based on GC-MS), assays containing internal controls will have to be devised. This will become essential to apply this tool clinically.
Previously, we reported that some metabolites vary with disease stage (Farshidfar et al, 2012). Indeed, we have replicated the metabolomic profile characterising metastatic CRC. In addition to reproducing our earlier observation, we demonstrated other metabolomic characteristics of CRC progression. Specifically, we identified a number of metabolomic features that accompanied lymphatic spread and also features that were associated with local tumour growth (as measured in terms of size or depth of invasion). These observations enabled exploration of some more defined applications of metabolomic biomarkers.
The identification of metabolic changes associated with nodal disease enabled an analysis of stage II tumours for possible occult nodal metastases. This was based on the observation that pathological nodal examination was an imperfect means of identifying nodal disease because of inadequate sampling by the surgeon or the pathologist (Cserni et al, 2002; Baxter et al, 2005; Doekhie et al, 2006). We were unable to identify individuals in the stage II cohort who had metabolomic features of stage III disease. This may be because the patients were accurately staged based on the traditional pathological examination (on average, 19.42±9.80 lymph nodes were examined), or because the metabolomic biomarker was not sufficiently sensitive to identify incorrectly staged individuals.
We also explored whether it was possible to prognosticate in stage II disease. This is important because it is known that some stage II CRCs recur early, and this has been attributed to the molecular features of these cancers (Zhou et al, 2002; Gray et al, 2011; Kennedy et al, 2011; Salazar et al, 2011). We were successful in identifying metabolomic features that characterised stage II tumours that recurred within 36 months. Further external validation will be required to determine whether this biomarker profile is independently predictive of survival. Moreover, the underlying biology related to this poor prognosis signature requires further interrogation. A convenient blood-based test that reliably prognosticates stage II patients may be useful for identifying a subgroup that would benefit from adjuvant chemotherapy.
Ideally, any biomarker would reflect some underlying biology. Using pathway analysis, we attempted to delineate how the diagnostic biomarker for CRC might inform our understanding of biology. While the pathways identified did reflect hallmarks of cancer (increased proliferation, decreased cell death, altered carbohydrate metabolism), the ability to extrapolate on tumour biology is limited when blood samples are used. Multiple processes contribute to the metabolomic profile of blood, including the effects of tumour, the response of the host to malignancy, effects of the gut microbiome and effects of other environmental exposures. Therefore, to generate truly informative information from metabolomics, separate analysis of tumour, host and microbiome will be required.
In conclusion, using GC-MS, we were able to identify and validate a diagnostic biomarker for CRC, and even adenomas were detectable in blood. Further testing in a screening population (where prevalence is much lower) will instruct on the utility of these diagnostic biomarkers for screening. We also have made some intriguing observations that point to future applications of related biomarkers for prognostication of stage II disease and for screening. These CRC biomarkers are poised for further development, which will entail external validation. Gas chromatography-mass spectrometry has substantial potential for application in a clinical laboratory because of its reproducibility and because GC-MS units are relatively compact. Importantly, because these biomarkers were all determined using a single analytical modality, it will be possible to devise a single assay that will simultaneously determine a range of diagnostic and prognostic biomarkers applicable to CRC. Future efforts will be directed at development of a quantitative assay, as well as further external validation using samples from multiple centres. In addition, further biomarker development will be important in hereditary forms of CRC, as well as CRC associated with inflammatory bowel disease.
Change history
27 September 2016
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
André T, Boni C, Navarro M, Tabernero J, Hickish T, Topham C, Bonetti A, Clingan P, Bridgewater J, Rivera F, de Gramont A (2009) Improved overall survival with oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment in stage II or III colon cancer in the MOSAIC Trial. J Clin Oncol 27 (19): 3109–3116.
Bathe O, Farshidfar F (2014) From genotype to functional phenotype: unraveling the metabolomic features of colorectal cancer. Genes 5 (3): 536–560.
Bathe OF, Shaykhutdinov R, Kopciuk K, Weljie AM, McKay A, Sutherland FR, Dixon E, Dunse N, Sotiropoulos D, Vogel HJ (2011) Feasibility of identifying pancreatic cancer based on serum metabolomics. Cancer Epidemiol Biomarkers Prev 20 (1): 140.
Baxter NN, Virnig DJ, Rothenberger DA, Morris AM, Jessurun J, Virnig BA (2005) Lymph node evaluation in colorectal cancer patients: a population-based study. J Natl Cancer Inst 97 (3): 219–225.
Bertini I, Cacciatore S, Jensen BV, Schou JV, Johansen JS, Kruhøffer M, Luchinat C, Nielsen DL, Turano P (2012) Metabolomic NMR fingerprinting to identify and predict survival of patients with metastatic colorectal cancer. Cancer Res 72 (1): 356–364.
Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37 (8): 911–917.
Croner RS, Geppert CI, Bader FG, Nitsche U, Spath C, Rosenberg R, Zettl A, Matias-Guiu X, Tarragona J, Guller U, Sturzl M, Zuber M (2014) Molecular staging of lymph node-negative colon carcinomas by one-step nucleic acid amplification (OSNA) results in upstaging of a quarter of patients in a prospective, European, multicentre study. Br J Cancer 110 (10): 2544–2550.
Cserni G, Vinh-Hung V, Burzykowski T (2002) Is there a minimum number of lymph nodes that should be histologically assessed for a reliable nodal staging of T3N0M0 colorectal carcinomas? J Surg Oncol 81 (2): 63–69.
Cubiella J, Salve M, Diaz-Ondina M, Vega P, Alves MT, Iglesias F, Sanchez E, Macia P, Blanco I, Bujanda L, Fernandez-Seara J (2014) Diagnostic accuracy of the faecal immunochemical test for colorectal cancer in symptomatic patients: comparison with NICE and SIGN referral criteria. Colorectal Dis 16 (8): O273–O282.
Dieterle F, Ross A, Schlotterbeck G, Senn H (2006) Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal Chem 78 (13): 4281–4290.
Doekhie FS, Kuppen PJ, Peeters KC, Mesker WE, van Soest RA, Morreau H, van de Velde CJ, Tanke HJ, Tollenaar RA (2006) Prognostic relevance of occult tumour cells in lymph nodes in colorectal cancer. Eur J Surg Oncol 32 (3): 253–258.
Dunn WB, Broadhurst DI, Atherton HJ, Goodacre R, Griffin JL (2011) Systems level studies of mammalian metabolomes: the roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chem Soc Rev 40 (1): 387–426.
Egdington ES (1987) Randomization Tests. Marcel Dekker Inc: New York, NY, USA.
Farshidfar F, Weljie AM, Kopciuk K, Buie WD, Maclean A, Dixon E, Sutherland FR, Molckovsky A, Vogel HJ, Bathe OF (2012) Serum metabolomic profile as a means to distinguish stage of colorectal cancer. Genome Med 4 (5): 42.
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5 (10): R80.
Gleisner AL, Mogal H, Dodson R, Efron J, Gearhart S, Wick E, Lidor A, Herman JM, Pawlik TM (2013) Nodal status, number of lymph nodes examined, and lymph node ratio: what defines prognosis after resection of colon adenocarcinoma? J Am Coll Surg 217 (6): 1090–1100.
Gray RG, Quirke P, Handley K, Lopatin M, Magill L, Baehner FL, Beaumont C, Clark-Langone KM, Yoshizawa CN, Lee M, Watson D, Shak S, Kerr DJ (2011) Validation study of a quantitative multigene reverse transcriptase-polymerase chain reaction assay for assessment of recurrence risk in patients with stage II colon cancer. J Clin Oncol 29 (35): 4611–4619.
Griffin JL, Nicholls AW (2006) Metabolomics as a functional genomic tool for understanding lipid dysfunction in diabetes, obesity and related disorders. Pharmacogenomics 7 (7): 1095–1107.
Heitman SJ, Hilsden RJ, Au F, Dowden S, Manns BJ (2010) Colorectal cancer screening for average-risk North Americans: An economic evaluation. PLoS Med 7 (11): e1000370.
Hollander M, Wolfe DA (1999) Nonparametric Statistical Methods 2nd edn Wiley: New York, NY, USA.
Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K, Oda Y, Kakazu Y, Kusano M, Tohge T, Matsuda F, Sawada Y, Hirai MY, Nakanishi H, Ikeda K, Akimoto N, Maoka T, Takahashi H, Ara T, Sakurai N, Suzuki H, Shibata D, Neumann S, Iida T, Tanaka K, Funatsu K, Matsuura F, Soga T, Taguchi R, Saito K, Nishioka T (2010) MassBank: a public repository for sharing mass spectral data for life sciences. J Mass Spectrom 45 (7): 703–714.
Hummel J, Selbig J, Walther D, Kopka J (2007) The Golm Metabolome Database: a database for GC-MS based metabolite profiling. In Metabolomics J Nielsen, M Jewett, (eds) Vol. 18, Chapter 229 pp 75–95. Springer: Berlin, Heidelberg, Germany.
Kanehisa M, Goto S (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28 (1): 27–30.
Kennedy RD, Bylesjo M, Kerr P, Davison T, Black JM, Kay EW, Holt RJ, Proutski V, Ahdesmaki M, Farztdinov V, Goffard N, Hey P, McDyer F, Mulligan K, Mussen J, O'Brien E, Oliver G, Walker SM, Mulligan JM, Wilson C, Winter A, O'Donoghue D, Mulcahy H, O'Sullivan J, Sheahan K, Hyland J, Dhir R, Bathe OF, Winqvist O, Manne U, Shanmugam C, Ramaswamy S, Leon EJ, Smith WI, McDermott U, Wilson RH, Longley D, Marshall J, Cummins R, Sargent DJ, Johnston PG, Harkin DP (2011) Development and independent validation of a prognostic assay for stage II colon cancer using formalin-fixed paraffin-embedded tissue. J Clin Oncol 29 (35): 4620–4626.
Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD (2012) The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28 (6): 882–883.
Leichtle AB, Nuoffer J-M, Ceglarek U, Kase J, Conrad T, Witzigmann H, Thiery J, Fiedler GM (2012) Serum amino acid profiles and their alterations in colorectal cancer. Metabolomics 8 (4): 643–653.
Ma Y, Zhang P, Wang F, Liu W, Yang J, Qin H (2012) An integrated proteomics and metabolomics approach for defining oncofetal biomarkers in the colorectal cancer. Ann Surg 255 (4): 720–730.
Mal M, Koh PK, Cheah PY, Chan ECY (2012) Metabotyping of human colorectal cancer using two-dimensional gas chromatography mass spectrometry. Anal Bioanal Chem 403 (2): 483–493.
Marisa L, de Reyniès A, Duval A, Selves J, Gaub MP, Vescovo L, Etienne-Grimaldi M-C, Schiappa R, Guenot D, Ayadi M, Kirzin S, Chazal M, Fléjou J-F, Benchimol D, Berger A, Lagarde A, Pencreach E, Piard F, Elias D, Parc Y, Olschwang S, Milano G, Laurent-Puig P, Boige V (2013) Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med 10 (5): e1001453.
Nishiumi S, Kobayashi T, Ikeda A, Yoshie T, Kibi M, Izumi Y, Okuno T, Hayashi N, Kawano S, Takenawa T, Azuma T, Yoshida M (2012) A novel serum metabolomics-based diagnostic approach for colorectal cancer. PLoS One 7 (7): e40459.
Qiu Y, Cai G, Su M, Chen T, Zheng X, Xu Y, Ni Y, Zhao A, Xu LX, Cai S, Jia W (2009) Serum metabolite profiling of human colorectal cancer using GC-TOFMS and UPLC-QTOFMS. J Proteome Res 8 (10): 4844–4850.
R Core Team (2014) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria.
Roth AD, Delorenzi M, Tejpar S, Yan P, Klingbiel D, Fiocca R, d’Ario G, Cisar L, Labianca R, Cunningham D, Nordlinger B, Bosman F, Van Cutsem E (2012) Integrated analysis of molecular and clinical prognostic factors in stage II/III colon cancer. J Natl Cancer Inst 104 (21): 1635–1646.
Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J (2003) TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34 (2): 374–378.
Salazar R, Roepman P, Capella G, Moreno V, Simon I, Dreezen C, Lopez-Doriga A, Santos C, Marijnen C, Westerga J, Bruin S, Kerr D, Kuppen P, van de Velde C, Morreau H, Van Velthuysen L, Glas AM, Van't Veer LJ, Tollenaar R (2010) Gene expression signature to improve prognosis prediction of stage II and III colorectal cancer. J Clin Oncol 29 (1): 17–24.
Salazar R, Roepman P, Capella G, Moreno V, Simon I, Dreezen C, Lopez-Doriga A, Santos C, Marijnen C, Westerga J, Bruin S, Kerr D, Kuppen P, van de Velde C, Morreau H, Van Velthuysen L, Glas AM, Van't Veer LJ, Tollenaar R (2011) Gene expression signature to improve prognosis prediction of stage II and III colorectal cancer. J Clin Oncol 29 (1): 17.
Sarli L, Bader G, Iusco D, Salvemini C, Mauro DD, Mazzeo A, Regina G, Roncoroni L (2005) Number of lymph nodes examined and prognosis of TNM stage II colorectal cancer. Eur J Cancer 41 (2): 272–279.
Slupsky CM, Rankin KN, Wagner J, Fu H, Chang D, Weljie AM, Saude EJ, Lix B, Adamko DJ, Shah S, Greiner R, Sykes BD, Marrie TJ (2007) Investigations of the effects of gender, diurnal variation, and age in human urinary metabolomic profiles. Anal Chem 79 (18): 6995–7004.
Smith RA, Manassaram-Baptiste D, Brooks D, Doroshenk M, Fedewa S, Saslow D, Brawley OW, Wender R (2015) Cancer screening in the United States, 2015: a review of current American Cancer Society guidelines and current issues in cancer screening. CA Cancer J Clin 65 (1): 30–54.
Stein S (1995) Chemical substructure identification by mass spectral library searching. J Am Soc Mass Spectrom 6 (8): 644–655.
Tan B, Qiu Y, Zou X, Chen T, Xie G, Cheng Y, Dong T, Zhao L, Feng B, Hu X, Xu LX, Zhao A, Zhang M, Cai G, Cai S, Zhou Z, Zheng M, Zhang Y, Jia W (2013) Metabonomics identifies serum metabolite markers of colorectal cancer. J Proteome Res 12 (6): 3000–3009.
The Cancer Genome Atlas Network (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature 487 (7407): 330–337.
Triba MN, Le Moyec L, Amathieu R, Goossens C, Bouchemal N, Nahon P, Rutledge DN, Savarin P (2015) PLS/OPLS models in metabolomics: the impact of permutation of dataset rows on the K-fold cross-validation quality parameters. Mol Biosyst 11 (1): 13–19.
UmetricsAB (2015) User Guide to SIMCA, Version 14. Umetrics AB: Malmö, Sweden.
Weljie AM, Dowlatabadi R, Miller BJ, Vogel HJ, Jirik FR (2007) An inflammatory arthritis-associated metabolite biomarker pattern revealed by 1H NMR spectroscopy. J Proteome Res 6 (9): 3456–3464.
Whitlock EP, Lin JS, Liles E, Beil TL, Fu R (2008) Screening for colorectal cancer: a targeted, updated systematic review for the US Preventive Services Task Force. Ann Intern Med 149 (9): 638–658.
Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, Fung C, Nikolai L, Lewis M, Coutouly M-A, Forsythe I, Tang P, Shrivastava S, Jeroncic K, Stothard P, Amegbey G, Block D, Hau DD, Wagner J, Miniaci J, Clements M, Gebremedhin M, Guo N, Zhang Y, Duggan GE, MacInnis GD, Weljie AM, Dowlatabadi R, Bamforth F, Clive D, Greiner R, Li L, Marrie T, Sykes BD, Vogel HJ, Querengesser L (2007) HMDB: the Human Metabolome Database. Nucleic Acids Res 35 (Suppl 1): D521–D526.
Zhou W, Goodman SN, Galizia G, Lieto E, Ferraraccio F, Pignatelli C, Purdie CA, Piris J, Morris R, Harrison DJ, Paty PB, Culliford A, Romans KE, Montgomery EA, Choti MA, Kinzler KW, Vogelstein B (2002) Counting alleles to predict recurrence of early-stage colorectal cancers. Lancet 359 (9302): 219–225.
Zhu J, Djukovic D, Deng L, Gu H, Himmati F, Chiorean EG, Raftery D (2014) Colorectal cancer detection using targeted serum metabolic profiling. J Proteome Res 13 (9): 4120–4130.
Acknowledgements
FF is the recipient of graduate scholarship from Alberta Cancer Foundation (ACF). HJV currently holds the Armstrong Chair in Molecular Cancer Epidemiology Research. We thank Nicole Dunse and Cameron Holder of the University of Calgary HPB/GI Tumour Bank and Dr Susanna Town of the Forzani and MacPhail Colon Cancer Screening Centre (CCSC) Biorepository for their invaluable contributions and support. We would also like to acknowledge the assistance of Dr Ruichuan Zhang, the mass spectrometry facility manager, for his continued support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
FF, AMW, KAK, HJV and OFB are coinventors of a related CRC diagnostic patent. AMW is funded for an unrelated project by Merck. RH is an advisor for Exact Sciences Inc., and is funded for an unrelated project by Advanced Proteomics Inc.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Farshidfar, F., Weljie, A., Kopciuk, K. et al. A validated metabolomic signature for colorectal cancer: exploration of the clinical value of metabolomics. Br J Cancer 115, 848–857 (2016). https://doi.org/10.1038/bjc.2016.243
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2016.243
Keywords
This article is cited by
-
Profiling the metabolic disorder and detection of colorectal cancer based on targeted amino acids metabolomics
Journal of Translational Medicine (2023)
-
Serum untargeted lipidomics by UHPLC-ESI-HRMS aids the biomarker discovery of colorectal adenoma
BMC Cancer (2022)
-
Plasma metabolomic profiles for colorectal cancer precursors in women
European Journal of Epidemiology (2022)
-
The role of gut microbiota in the development of colorectal cancer: a review
International Journal of Colorectal Disease (2022)
-
Metabolic signatures of greater body size and their associations with risk of colorectal and endometrial cancers in the European Prospective Investigation into Cancer and Nutrition
BMC Medicine (2021)