Abstract
Poly-γ-glutamic acid (γ-PGA) is an important biochemical product with a variety of applications. This work reports a novel approach to improve γ-PGA through over expression of key enzymes in cofactor NADPH generating process for NADPH pool. Six genes encoding the key enzymes in NADPH generation were over-expressed in the γ-PGA producing strain B. licheniformis WX-02. Among various recombinants, the strain over-expressing zwf gene (coding for glucose-6-phosphate dehydrogenase), WX-zwf, produced the highest γ-PGA concentration (9.13 g/L), 35% improvement compared to the control strain WX-pHY300. However, the growth rates and glucose uptake rates of the mutant WX-zwf were decreased. The transcriptional levels of the genes pgsB and pgsC responsible for γ-PGA biosynthesis were increased by 8.21- and 5.26-fold, respectively. The Zwf activity of the zwf over expression strain increased by 9.28-fold, which led to the improvement of the NADPH generation, and decrease of accumulation of by-products acetoin and 2,3-butanediol. Collectively, these results demonstrated that NADPH generation via over-expression of Zwf is as an effective strategy to improve the γ-PGA production in B. licheniformis.
Similar content being viewed by others
Introduction
Poly-γ-glutamic acid (γ-PGA) is a multifunctional biopolymer consisting of D- and L-glutamate as monomers. Due to its features of cation chelating, hygroscopicity, water-solubility, non-toxicity and biodegradability, γ-PGA has been applied in cosmetics, medicine, food, water treatment, and agricultural industries, such as material and drug carrier, food preservative, metal ion chelating agents and highly water absorbable hydrogels1,2,3.
γ-PGA is commonly produced through bacterial transformation of glutamate. In general, Bacillus strains were the major γ-PGA producers1,4. Among Bacillus strains, B. licheniformis WX-02 has been proven as a promising γ-PGA producer5. Some researches have been performed to improve γ-PGA production by this strain. For example, nitrate addition improved γ-PGA by 2.3-fold6, and the physicochemical stresses such as heat, osmotic and alkaline improved the γ-PGA yield7,8. Also, over-expression of PgdS (γ-PGA hydrolase) and Glr (glutamic acid racemase) could enhance γ-PGA production9,10. Meanwhile, the research conducted in our group has revealed that glutamate dehydrogenase RocG played a critical role for the conversion of α-ketoglutarate to glutamic acid, and the function of RocG from B. licheniformis WX-02 was different from that of B. subtilis11. Also, RocG of B. licheniformis was confirmed as a NADPH-dependent enzyme during the γ-PGA production12.
In recent years, improvement of γ-PGA production in Bacillus species has been focused on engineering of the strains’ metabolic pathway4. Glutamic acid is the precursor for the synthesis of γ-PGA, and it is mainly converted by α-ketoglutarate in TCA cycle when no glutamic acid is added into the medium. Also, researchers have identified the pgsB, pgsC and pgsA as essential genes in the γ-PGA biosynthesis, and gene degU and swrA were reported as positive regulators for the expression of PgsBCA13. Based on this basic information, various strategies have been developed to manipulate the metabolic pathway for improving γ-PGA production. For example, genes involving in the byproduct biosynthesis (sac responsible for levan synthesis, lps responsible for lipopolysaccharide synthesis) were knocked out in Bacillus amyloliquefaciens LL314,15. Also, Scoffone et al., deleted the genes coding for the major γ-PGA degrading enzymes, pgdS and ggt to enhance the γ-PGA production, and the γ-PGA purity and yield were all increased16.
Regulation of cofactor such as NADH and NADPH is another potential method to enhance product production17. In general, NADH and NADPH serve as cofactors in various metabolic pathways in bacterial cells. The cofactor NADH plays an important role in the catabolism, and NADPH acts as an essential component for the anabolism. This cofactor has proved as an important cofactor in the synthesis of biopolymers18,19. NADPH is produced in pentose phosphate pathway (PPP), tricarboxylic acid cycle (TCA) and transhydrogenases system20. Xu et al. over-expressed NAD kinase in Corynebacterium crenatum to improve polyhydroxybutyrate (PHB) yield by 15.7%21. In E. coli, over-expression of NADPH transhydrogenase UdhA was used as an effective way to improve PHB production22. Lim et al. improved the PHB production by 41% in the Zwf over-expressed strain23.
Based on the previous research, we explore the strategy of cofactor regulation to improve γ-PGA production. Here, we hypothesize that the γ-PGA synthesis of B. licheniformis WX-02 could be improved by over-expressing genes related with NADPH generation such as gnd, zwf, gdh and ppnk from B. licheniformis WX-02, pntAB and udhA from E. coli BL21(DE3). The aim of this work is to improve the γ-PGA production by cofactor NADPH generation. The research provides valuable information on a new approach to improve γ-PGA production.
Results
Establishment of the recombinant strains
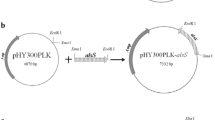
Six enzymes (Zwf, Gnd, Gdh, Ppnk, PntAB and UdhA) related with NADPH production were respectively over-expressed in B. licheniformis WX-02 to improve the γ-PGA production (Fig. 1). Here, the construction of expression plasmid pHY-zwf was used as an example (Fig. S1). The plasmid pHY-zwf harboring the P43 promoter (K02174.1), target gene zwf (FJ374767.1) and amyL terminator (FJ556804.1) was confirmed through PCR verification and DNA sequencing. The plasmid pHY-zwf was then electro-transferred into the native strain B. licheniformis WX-02 to construct the recombinant strain B. licheniformis WX-zwf. Other recombinants were constructed using the same method and were named WX-gnd, WX-gdh, WX-ppnk, WX-pntAB and WX-udhA, respectively. B. licheniformis WX-02 and WX-pHY300 were served as the control strains.
Production of γ-PGA by the NADPH over-expression strains
Six recombinant strains (WX-zwf, WX-gnd, WX-gdh, WX-ppnk, WX-pntAB and WX-udhA) and two control strains (WX-02, WX-pHY300) were cultured in γ-PGA production medium to evaluate the effects of NADPH generation on the γ-PGA production. As shown in Fig. 2, WX-zwf showed the highest γ-PGA yield (9.13 g/L) among five recombinant strains, and was 35% higher than that of the control WX-pHY300. The γ-PGA yields of other recombinant strains WX-udhA (8.24 g/L), WX-pntAB (8.01 g/L), WX-gnd (7.55 g/L) and WX-ppnk (7.35 g/L) were 22%, 19%, 12% and 9% higher than that of the control strain WX-pHY300. While the strain WX-gdh did not improve γ-PGA produciton compared to the control strain WX-pHY300. The results indicate that over-expression of the enzymes Zwf, UdhA, PntAB, Gnd and Ppnk in NADPH generation is an effective way to improve γ-PGA production, among which Zwf exhibited the highest improvement of γ-PGA production.
The fermentation of WX-pHY300 and WX-zwf were further performed to determine the time course of γ-PGA production, cell growth, residual glucose, and main byproducts (acetoin and 2,3-butanediol) produced (Fig. 3). As shown in Fig. 3, the control strain WX-pHY300 rapidly entered into the exponential phase after 8-h of lag phase, γ-PGA synthesized rapidly along with rapid cell growth and glucose consumption, these trends were similar to those in previous reports8. Compared to the control strain WX-pHY300, the mutant strain WX-zwf had a much longer lag phase (18 h) with a slow growth rate. However, the γ-PGA yield by the WX-zwf strain increased 35%, and the cellular γ-PGA content improved from 1.32 g/gDCW to 1.72 g/gDCW in compared that of the WX-pHY300. The γ-PGA produced by glucose was 0.15 g/gglc, 35% increase compared with that of WX-pHY300 (0.11 g/gglc). Since WX-zwf had a much longer lag phase (18 h) with a slow growth rate, the production rate of WX-zwf was 0.21 g/L/hr, which has no difference compared with that of WX-pHY300 (0.21 g/L/hr). Also, the glucose uptake rate of WX-zwf was 0.23 g/gDCW·h, 36% lower than that of WX-pHY300. Meanwhile, the production of byproducts (acetoin and 2,3-butanediol) decreased 18% in the WX-zwf.
Transcriptional level of Zwf over-expression strain
The transcriptional levels of the genes involved in the γ-PGA biosynthesis and glucose metabolism were determined. As shown in Fig. 4A, the zwf transcriptional level in WX-zwf strain was increased by 45.96-fold compared to the control strain WX-pHY300. The gnd transcriptional level was also increased (by 3.03-fold) due to the over-expression of Zwf, which might promote NADPH generation as well. Also, 2-dehydro-3-deoxy-phosphogluconate aldolase gene eda and phosphogluconate dehydratase gene edd in the ED pathway were increased by 0.45-fold and 0.37-fold, and the increase rates were much low than that of gnd. Previously, other report using 13C labeling indicated that redirecting carbon fluxes to the oxidative pentose phosphate pathway may lead to minor increases of the ED pathway fluxes, which was positively correlated with our results24. The transcriptional level of glucose-6-phosphate isomerase gene pgi and glyceraldehyde-3-phosphate dehydrogenase gene gapA in the glycolysis pathway were decreased because of the strengthening of PPP, while the citrate synthase gene citB and isocitrate dehydrogenase gene icd transcriptional levels were increased. Meanwhile, acted as the γ-PGA biosynthesis genes, the pgsB and pgsC transcriptional levels were increased by 8.21-fold and 5.26-fold, respectively. As the essential regulators to activate the γ-PGA biosynthesis, the transcriptional levels of degU and swrA were up-regulated by 3.45-fold and 3.21-fold (Fig. 4B), which might improve the expression of γ-PGA biosynthetase PgsABC13. Also, the improvement of the anaerobic regulator gene fnr transcriptional might improve the nitrate reduction and ATP supplement for γ-PGA production25. Furthermore, the genes (ackA, adhE and ldh) responsible for acetate, ethanol and lactate synthesis were all decreased in the Zwf overexpression strain, which indicated that over-expression of Zwf could affect the overflow metabolism.
Zwf activity of Zwf over-expression strains
The Zwf activity was determined every 4 hours during the γ-PGA production stage. As shown in Fig. 5, Zwf was expressed along with the rapid cell growth. The highest Zwf activity of control strain WX-pHY300 (852 U/gDCW) was achieved at the mid-log phase (16 h). The Zwf activity of the recombinant strain WX-zwf was higher than that of WX-pHY300 throughout the fermentation process, with the highest Zwf activity reaching 8762 U/gDCW at 36 h, a 9.28-fold higher than WX-pHY300 strain. The Zwf activity was decreased significantly at the stationary phase (Fig. 5).
NADPH and NADH production of Zwf over-expression strain
NADPH level was an important factor for γ-PGA biosynthesis, and the highest NADPH content was attained at the exponential phase in each strain, WX-zwf produce 34.49 μmol/gDCW NADPH, 2.36-fold higher than that of WX-pHY300. Meanwhile NADH content in WX-zwf decreased by 27% compared to the control pHY300 strain (Table 1). The ratio of NADPH/NADH was 2.99, a 3.60-fold higher than that of WX-pHY300. Collectively, the results indicated that Zwf over-expression improved the NADPH production, which in turn promoted the glutamic acid and γ-PGA biosynthesis. Meanwhile, NADH plays the vital role in the biosynthesis of 2,3-butanediol26,27. A decrease of NADH pool in the WX-zwf strains might be reason for the decline of acetoin/2,3-butanediol production.
Discussion
γ-PGA is an important product with many applications1. Metabolic engineering is an effective strategy for the improving the synthesis of target products including γ-PGA. Many tactics have been developed to improve γ-PGA production, including blocking the by-products synthesis pathways and enhancing the glutamic acid synthesis pathways4,14. However, the strategy of regulating cofactor such as NADPH for improving γ-PGA production has not been attempted, although cofactor regulation in general has been recognized as an effective method for product synthesis. In this report, it was confirmed that NADPH generation was a limiting factor in γ-PGA biosynthesis, over-expression of Zwf was an effective way to improve the γ-PGA production.
Six recombinant strains were constructed to improve the γ-PGA production by NADPH generation, and the γ-PGA yield was in a descending order for those strains as WX-zwf > WX-udhA > WX-pntAB > WX-gnd > WX-ppnk > WX-gdh > WX-pHY300 > WX-02. Over-expression of Zwf showed the most promising outcome. A 12% improvement was obtained in the Gnd over-expressed strain WX-gnd, which was much lower than that of WX-zwf. Since glucose can barely enter cells by Gdh, over-expression of Gdh is not effective to improve NADPH generation and γ-PGA yield. These results were in agreement with the previous research23. It should be noted that the Zwf over-expressed strain had a low glucose consumption rate, cell growth rate and γ-PGA biosynthesis rate at the initial phase, there were more rooms to optimize this mutant strains by genetic modification and process engineering.
Among PPP, TCA cycle and the transhydrogenases system pathways, PPP pathway contributes most to NADPH generation. As the first and key gene in PPP, Zwf converts glucose-6-phosphate to 6-phosphogluconate with the conversion of NADP+ to NADPH20, therefore, over-expression of Zwf increases the NADPH generation, which in turn accelerate the reaction of α-ketoglutarate to glutamate and finally improve the γ-PGA production. Meanwhile, over-expression of Zwf could mainly strengthen the OPP pathway, and strengthening of OPP pathway could promote NADPH generation. Meanwhile, Over-expression of Zwf also weakens the glycolysis pathway, which then reduces the overflow metabolism and byproduct synthesis. Also, the TCA cycle was strengthened in the Zwf over-expression strain, which was beneficial for the γ-PGA production. Collectively, this study shows that the over-expression of Zwf is an effective strategy to improve the yield of target product such as γ-PGA in the NADPH-dependent biotransformation pathway, compared with over-expression of other genes.
Over-expression of Zwf could weaken the glycolysis pathway, which might affect the overflow metabolism. Also, Over-expression of Zwf could convert more NADH and NADP+ to NAD+ and NADPH respectively, which will decrease the NADH accumulation in the Zwf over-expression strain. Previous reports have proved that NADH plays an important role in the biosynthesis of the acetoin and 2,3-butanediol26. The yields of the acetoin and 2,3-butanediol were decreased due to the reduction of the NADH concentration in the strain WX-zwf. Collectively, the increase of the γ-PGA yield was considered due to the improvement of NADPH generation via Zwf over-expression, and the decrease in the synthesis of acetoin/2,3-butanediol byproducts resulted from the reduction of NADH pool.
Served as the two important transcriptional regulation gene for γ-PGA biosynthesis, the increase of expression levels of degU and swrA could activate the expression of γ-PGA synthetase13. Also, Fnr was served as the main anaerobic regulator in Bacillus, and the increase of fnr transcriptional level might improve the nitrate respiration and ATP synthesis25. Our results also indicate that NADPH generation might improve the quorum sensing system and anaerobic metabolism in B. licheniformis, which improve the transcriptional levels of pgsB and γ-PGA production.
Conclusion
In this study, six enzymes were respectively over-expressed to improve the γ-PGA production in B. licheniformis WX-02. Among the mutant strains, rhe Zwf over-expressed strain WX-zwf showed the highest yield of γ-PGA (9.13 g/L), 35% higher than that of the control strina WX-pHY300. Also, the transcriptional levels of genes responsible for γ-PGA synthesis and Zwf activity were correlated positively with γ-PGA production. The over-expression of Zwf also increased the NADPH concentration as well as the ratio of NADPH/NADH, which led to the improvement of γ-PGA production. The concentration of the main byproduct acetoin/2,3-butanediol was decreased obviously due to the reduction of NADH concentration. Collectively, the results demonstrated that enhanced NADPH generation via over-expression of Zwf can be as an effective strategy to improve the γ-PGA production.
Materials and Methods
Bacterial strains and plasmids
The bacterial strains and plasmids used in this study are listed in Table 2. B. licheniformis WX-02 (CCTCC M208065) was served as the native strain for the construction of recombinant strains5. E. coli DH5α was the host strain for plasmid construction. The plasmid pHY300PLK was used as the expression vector28.
Media and culture conditions
LB medium containing (per liter) 10 g peptone, 5 g yeast extract, and 10 g NaCl (pH 7.2) was used as the basic medium. When necessary, ampicillin (50 mg/L) was added to the E. coli culture, while tetracycline (20 mg/L) was added to the B. licheniformis culture. The seed culture of B. licheniformis was prepared in 250 mL flasks containing 50 mL LB medium and incubated at 37 °C in a rotatory shaker (180 rpm) for 10 h until OD600 reach 4.0~4.5. The seed was then transferred (3% inoculum ratio) to γ-PGA production medium containing (per liter) 60 g glucose, 10 g sodium nitrate, 10 g sodium citrate, 8 g NH4Cl, 1 g CaCl2, 1 g K2HPO4·3H2O, 1 g MgSO4·7H2O, 1 g ZnSO4·7H2O and 0.15 g MnSO4·7H2O at pH 7.26. The incubating conditions were the same as those of the seed culture. All the fermentation experiments were performed in three replicates.
Construction of the over-expression strains
The expression vectors were constructed based on pHY300PLK. The construction of the plasmid containing different genes was exemplified with pHY-zwf (Fig. S1)28. Firstly, P43 promoter (K02174.1) from B. subtilis 168, the zwf gene (52783855) and amyL terminator (FJ556804.1) from B. licheniformis WX-02 were amplified with the corresponding primers (Table 3). The amplified fragments were ligated by Splicing Overlapping Extension PCR (SOE-PCR) with the primers of P43-F and TamyL-R, and then cloned into pHY300PLK at the restriction sites EcoRI and XbaI to form the Zwf over-expressing vector, named pHY-zwf. The pHY-zwf vector was then transferred into B. licheniformis WX-02 based on the method reported previously29. The recombinant strain WX-zwf was confirmed by PCR and plasmids extraction.
The strains over-expressing Gdh, Gnd, Ppnk, PntAB and UdhA were constructed using the same method as that in Zwf, and were respectively named as B. licheniformis WX-gdh, WX-gnd, WX-ppnk, WX-pntAB and WX-udhA. The vector pHY300PLK were also transformed into native WX-02 strain to form the control strain WX-pHY300.
Determination of γ-PGA production, biomass and residue glucose
γ-PGA was determined by HPLC based on the method reported previously8. A 7.8 mm × 300 mm TSK Gel G6000 PWXL gel permeation chromatogram column (Tosoh, Tokyo, Japan) was used. The samples were eluted with a mixture of 25 mM sodium sulfate solution: acetonitrile (8:1) with the flow rate of 0.5 mL/min and detected at 220 nm. The γ-PGA yield was quantified based on the standard curve. To determine the cell dry weight, 6 mL distilled water was mixed with 2 mL broth, and the pH of the mixture was adjusted to 2.5 by 6 M HCl. The resulting solution was centrifuged at 12,000 g for 10 min, the cell pellets were harvested and dried at 80 °C. The residual glucose in the medium was detected by the SBA-40C bioanalyzer (Academy of science, Shandong, China).
Determination of acetoin and 2,3-butanediol concentrations
Acetoin and 2,3-butanediol were determined according to the methods reported previously30,31. In brief, the fermentation broth was mixed with equal volume of ethanol (containing 10 g/L butyl alcohol), centrifuged at 10,000 g for 15 min and filtrated through 0.22 μm filter. An Agilent 7890A gas chromatography with the flame ionization detector was then used to identify and quantify acetoin and 2,3-butanediol based on internal standard.
Analysis of transcription level
Total RNA was extracted by TRIzol® Reagent (Invitrogen, USA) according to previous method32. Trace DNA was digested by the Rnase-free DNase I enzyme (TaKaRa, Japan), and RevertAid First Strand cDNA Synthesis Kit (Thermo, USA) was applied to amplify the first stand of cDNA. The primers in Table S1 (seeing Supplementary Material) were used for amplifying the corresponding genes, and 16S rRNA from B. licheniformis WX-02 strain was used as the reference gene to normalize the data. The transcriptional levels for genes in the recombinant strain were compared with those of control strain after normalization to the reference gene 16S rRNA. All the experiments were performed in triplicate.
Assay of the Zwf
The Zwf activity was determined based on the previous method with minor modifications33. In brief, the cells were washed three times with cold disruption buffer (100 mM Tris-HCl, 20 mM KCl, 10 mM MgCl2, pH 7.5), followed with sonication (pulse: 1 s on; 2 s off; total 4 min) to disrupt cells. The solution with disrupted cells was centrifuged at 12,000 g for 20 min. The Zwf activity was determined containing 100 mM Tris-HCl (pH 7.5), 20 mM KCl, 1 mM NADP+, 10 mM MgCl2 and 5 mM Glucose-6-phosphate. The volume of 2.85 mL mixture was incubated at 30 °C for 5 min, and 150 μL samples were added to start the reactions, and the slope of the A340 at the initial reaction (for 0 to 4 min) was calculated, The Zwf activity was calculated as follows, where ΔA was the change of A340 per min, V is the reaction volume, ε is the molar extinction coefficient of the NADPH (6.22 × 103 L/(mol·cm)), and b is the light pathlength of the cuvette.

Determination of NADH and NADPH contents
NADH and NADPH contents were determined by previous method with minor modifications34,35. For NADH, 20 μL disrupted cell solution were added into plate wells containing 0.1 mL of 0.1 M HEPES (pH 7.5) and 2 mM EDTA, 20 μL of 1.2 mM DCPIP, 10 μL of 20 mM PMS, 25 μL water and 10 μL alcohol dehydrogenase (containing in 0.1 M HEPES (pH 7.5) and 2 mM EDTA). Ethanol (15 μL) was then added to the wells to start the reaction. The decrease of A600 was monitored every 30 s for 5 min. The contents were calculated by reference to the standards run concurrently (0~40 pmol NADH in the well). To determine NADPH, G6PDH was dissolved in 0.1 M HEPES (pH 7.5) and 2 mM EDTA, and 30 μL neutralized supernatant were introduced into the plates wells containing 0.1 mL of 0.1 M HEPES (pH 7.5), 2 mM EDTA, 20 μL of 1.2 mM DCPIP, 10 μL of 20 mM PMS, 30 μL water. The reaction was started by the addition of 10 μL G6PDH; the decrease in A600 was monitored every 30 s for 5 min. The rates were calculated with the standards, similar to the NADH analysis.
Additional Information
How to cite this article: Cai, D. et al. A novel approach to improve poly-γ-glutamic acid production by NADPH Regeneration in Bacillus licheniformis WX-02. Sci. Rep. 7, 43404; doi: 10.1038/srep43404 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Candela, T. & Fouet, A. Poly-gamma-glutamate in bacteria. Mol Microbiol 60, 1091–1098, doi: 10.1111/j.1365-2958.2006.05179.x (2006).
Lee, N. R. et al. In vitro evaluation of new functional properties of poly-gamma-glutamic acid produced by Bacillus subtilis D7. Saudi J Biol Sci 21, 153–158, doi: 10.1016/j.sjbs.2013.09.004 (2014).
Ogunleye, A. et al. Poly-gamma-glutamic acid: production, properties and applications. Microbiology 161, 1–17, doi: 10.1099/mic.0.081448-0 (2015).
Luo, Z. et al. Microbial synthesis of poly-gamma-glutamic acid: current progress, challenges, and future perspectives. Biotechnol Biofuels 9, 134, doi: 10.1186/s13068-016-0537-7 (2016).
Wei, X., Ji, Z. & Chen, S. Isolation of halotolerant Bacillus licheniformis WX-02 and regulatory effects of sodium chloride on yield and molecular sizes of poly-gamma-glutamic acid. Appl Biochem Biotechnol 160, 1332–1340, doi: 10.1007/s12010-009-8681-1 (2010).
Li, X. et al. Physiological and metabolic analysis of nitrate reduction on poly-gamma-glutamic acid synthesis in Bacillus licheniformis WX-02. Arch Microbiol 196, 791–799, doi: 10.1007/s00203-014-1014-y (2014).
Wei, X., Tian, G., Ji, Z. & Chen, S. A new strategy for enhancement of poly-γ-glutamic acid production by multiple physicochemical stresses in Bacillus licheniformis WX-02. J Chem Technol Biotechnol 90, 709–713, doi: 10.1128/JB.186.11.3399-3407.2004 (2015).
Wang, J., Yuan, H., Wei, X., Chen, J. & Chen, S. Enhancement of poly-γ-glutamic acid production by alkaline pH stress treatment in Bacillus licheniformis WX-02. J Chem Technol Biotechnol 121, 1444–1447 (2015).
Tian, G. et al. Enhanced expression of pgdS, gene for high production of poly-γ-glutamic aicd with lower molecular weight in Bacillus licheniformis WX-02. J Chem Technol Biotechnol 89, 1825–1832, doi: 10.1038/srep01377 (2013).
Jiang, F. et al. Expression of glr gene encoding glutamate racemase in Bacillus licheniformis WX-02 and its regulatory effects on synthesis of poly-gamma-glutamic acid. Biotechnol Lett 33, 1837–1840, doi: 10.1007/s10529-011-0631-7 (2011).
Gunka, K. & Commichau, F. M. Control of glutamate homeostasis in Bacillus subtilis: a complex interplay between ammonium assimilation, glutamate biosynthesis and degradation. Mol Microbiol 85, 213–224, doi: 10.1111/j.1365-2958.2012.08105.x (2012).
Tian, G., Wang, Q., Wei, X., Ma, X. & Chen, S. Glutamate dehydrogenase (RocG) in Bacillus licheniformis WX-02: enzymatic properties and specific functions in glutamic acid synthesis for Poly-γ-glutamic acid production. Enzyme Microb Technol 99, 9–15, doi: 10.1016/j.enzmictec.2017.01.002 (2017).
Ohsawa, T., Tsukahara, K. & Ogura, M. Bacillus subtilis response regulator DegU is a direct activator of pgsB transcription involved in gamma-poly-glutamic acid synthesis. Biosci Biotechnol Biochem 73, 2096–2102, doi: 10.1271/bbb.90341 (2009).
Feng, J. et al. Improved poly-gamma-glutamic acid production in Bacillus amyloliquefaciens by modular pathway engineering. Metab Eng 32, 106–115, doi: 10.1016/j.ymben.2015.09.011 (2015).
Zhang, W. et al. Deletion of genes involved in glutamate metabolism to improve poly-gamma-glutamic acid production in B. amyloliquefaciens LL3. J Ind Microbiol Biotechnol 42, 297–305, doi: 10.1007/s10295-014-1563-8 (2015).
Scoffone, V. et al. Knockout of pgdS and ggt genes improves gamma-PGA yield in B. subtilis . Biotechnol Bioeng 110, 2006–2012, doi: 10.1002/bit.24846 (2013).
Chemler, J. A., Fowler, Z. L., McHugh, K. P. & Koffas, M. A. Improving NADPH availability for natural product biosynthesis in Escherichia coli by metabolic engineering. Metab Eng 12, 96–104, doi: 10.1016/j.ymben.2009.07.003 (2010).
Lee, J. N., Shin, H. D. & Lee, Y. H. Metabolic engineering of pentose phosphate pathway in Ralstoniaeutropha for enhanced biosynthesis of poly-beta-hydroxybutyrate. Biotechnol Prog 19, 1444–1449, doi: 10.1021/bp034060v (2003).
Zhou, J., Liu, L., Shi, Z., Du, G. & Chen, J. ATP in current biotechnology: regulation, applications and perspectives. Biotechnol Adv 27, 94–101, doi: 10.1016/j.biotechadv.2008.10.005 (2009).
Lee, W. H., Kim, M. D., Jin, Y. S. & Seo, J. H. Engineering of NADPH regenerators in Escherichia coli for enhanced biotransformation. Appl Microbiol Biotechnol 97, 2761–2772, doi: 10.1007/s00253-013-4750-z (2013).
Xu, M. et al. Effect of Polyhydroxybutyrate (PHB) storage on L-arginine production in recombinant Corynebacterium crenatum using coenzyme regulation. Microb Cell Fact 15, 15, doi: 10.1186/s12934-016-0414-x (2016).
Sanchez, A. M., Andrews, J., Hussein, I., Bennett, G. N. & San, K. Y. Effect of overexpression of a soluble pyridine nucleotide transhydrogenase (UdhA) on the production of poly(3-hydroxybutyrate) in Escherichia coli . Biotechnol Prog 22, 420–425, doi: 10.1021/bp050375u (2006).
Lim, S. J., Jung, Y. M., Shin, H. D. & Lee, Y. H. Amplification of the NADPH-related genes zwf and gnd for the oddball biosynthesis of PHB in an E. coli transformant harboring a cloned phbCAB operon. J Biosci Bioeng 93, 543–549 (2002).
Hollinshead, W. D. et al. Examining Escherichia coli glycolytic pathways, catabolite repression, and metabolite channeling using Δpfk mutants. Biotechnol Biofuels 9, 212, doi: 10.1186/s13068-016-0630-y (2016).
Hartig, E. & Jahn, D. Regulation of the anaerobic metabolism in Bacillus subtilis . Adv Microb Phys 61, 195–216, doi: 10.1016/B978-0-12-394423-8.00005-6 (2012).
Fu, J. et al. NADH plays the vital role for chiral pure D-(−)-2,3-butanediol production in Bacillus subtilis under limited oxygen conditions. Biotechnol Bioeng 111, 2126–2131, doi: 10.1002/bit.25265 (2014).
Fu, J. et al. Metabolic engineering of Bacillus subtilis for chiral pure meso-2,3-butanediol production. Biotechnol Biofuels 9, 90, doi: 10.1186/s13068-016-0502-5 (2016).
Cai, D. et al. High-level expression of nattokinase in Bacillus licheniformis by manipulating signal peptide and signal peptidase. J Appl Microbiol 121, 704–712, doi: 10.1111/jam.13175 (2016).
Xue, G. P., Johnson, J. S. & Dalrymple, B. P. High osmolarity improves the electro-transformation efficiency of the gram-positive bacteria Bacillus subtilis and Bacillus licheniformis . J Microbiol Meth 34, 183–191 (1999).
Qiu, Y. et al. Engineering Bacillus licheniformis for the production of meso-2,3-butanediol. Biotechnol Biofuels 9, 117, doi: 10.1186/s13068-016-0522-1 (2016).
Qi, G. et al. Deletion of meso-2,3-butanediol dehydrogenase gene budC for enhanced D-2,3-butanediol production in Bacillus licheniformis . Biotechnol Biofuels 7, 16, doi: 10.1186/1754-6834-7-16 (2014).
Qiu, Y., Xiao, F., Wei, X., Wen, Z. & Chen, S. Improvement of lichenysin production in Bacillus licheniformis by replacement of native promoter of lichenysin biosynthesis operon and medium optimization. Appl Microbiol Biotechnol 98, 8895–8903, doi: 10.1007/s00253-014-5978-y (2014).
Wang, Z., Chen, T., Ma, X., Shen, Z. & Zhao, X. Enhancement of riboflavin production with Bacillus subtilis by expression and site-directed mutagenesis of zwf and gnd gene from Corynebacterium glutamicum . Bioresour Technol 102, 3934–3940, doi: 10.1016/j.biortech.2010.11.120 (2011).
Queval, G. & Noctor, G. A plate reader method for the measurement of NAD, NADP, glutathione, and ascorbate in tissue extracts: Application to redox profiling during Arabidopsis rosette development. Anal Biochem 363, 58–69, doi: 10.1016/j.ab.2007.01.005 (2007).
Tang, Z. et al. Improved oxytetracycline production in Streptomyces rimosus M4018 by metabolic engineering of the G6PDH gene in the pentose phosphate pathway. Enzyme Microb Technol 49, 17–24, doi: 10.1016/j.enzmictec.2011.04.002 (2011).
Wei, X. et al. Efficient expression of nattokinase in Bacillus licheniformis: host strain construction and signal peptide optimization. J Ind Microbiol Biotechnol 42, 287–295, doi: 10.1007/s10295-014-1559-4 (2015).
Acknowledgements
This work was supported by the National Science & Technology Pillar Program during the Twelfth Five-year Plan Period (2013AA102801-52), the National Program on Key Basic Research Project (973 Program, No. 2015CB150505), the Science and Technology Program of Wuhan (20160201010086).
Author information
Authors and Affiliations
Contributions
D. Cai and S. Chen designed the study. D. Cai carried out the molecular biology studies and construction of engineering strains. D. Cai, P. He, X. Lu and J. Zhu carried out the fermentation studies. D. Cai, C. Zhu, Y. Zhan, Q. Wang, Z. Wen and S. Chen analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Cai, D., He, P., Lu, X. et al. A novel approach to improve poly-γ-glutamic acid production by NADPH Regeneration in Bacillus licheniformis WX-02. Sci Rep 7, 43404 (2017). https://doi.org/10.1038/srep43404
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep43404
This article is cited by
-
Optimization of trans-4-hydroxyproline synthesis pathway by rearrangement center carbon metabolism in Escherichia coli
Microbial Cell Factories (2023)
-
Engineering of global transcription factors in Bacillus, a genetic tool for increasing product yields: a bioprocess overview
World Journal of Microbiology and Biotechnology (2023)
-
Construct a synthetic Entner–Doudoroff pathway in Bacillus licheniformis for enhancing lichenysin production
World Journal of Microbiology and Biotechnology (2023)
-
Poly-gamma-glutamic acid biopolymer: a sleeping giant with diverse applications and unique opportunities for commercialization
Biomass Conversion and Biorefinery (2023)
-
Engineering of a newly isolated Bacillus tequilensis BL01 for poly-γ-glutamic acid production from citric acid
Microbial Cell Factories (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.