Abstract
The anti-cancer activity of the benzo[h]quinolines was evaluated on cultured human skin cancer (G361), lung cancer (H460), breast cancer (MCF7) and colon cancer (HCT116) cell lines. The inhibitory effect of these compounds on the cell growth was determined by the MTT assay. The compounds 3e, 3f, 3h and 3j showed potential cytotoxicity against these human cancer cell lines. Effect of active compounds on DNA oxidation and expression of apoptosis related gene was studied. We also developed a quantitative method to measure the activity of cyclin-dependent kinases-2 (CDK2) by western blotting in the presence of active compound. In addition, molecular docking revealed that benzo[h]quinolines can correctly dock into the hydrophobic pocket of the targets receptor protein aromatase and CDK2, while their bioavailability/drug-likeness was predicted to be acceptable but requires future optimization. These findings reveal that benzo[h]quinolines act as anti-cancer agents by inducing oxidative stress-mediated DNA damage.
Similar content being viewed by others
Introduction
Cancer remains a major health problem worldwide with significant morbidity and mortality rates, causing about 14.6% of all worldwide deaths1. Currently, radio and chemotherapy are used for cancer treatment. In many cases these are successful, while in some cases they lead to severe compromising of the immune system due to lack of cell selectivity2. The treatment of cancer is indeed associated with many side effects, which include mutagenicity, tumorigenicity, skin irritation, developmental toxicity, hepatotoxicity, drug-induced cancer, alopecia etc resulting in a strong need for novel anti-cancer agents. In the last decade, several quinoline analogues have been shown to have anti-cancer activity. The quinoline ring system and its fused derivatives are significant structural units and are found in various alkaloids, therapeutics and synthetic analogues, which exhibit diverse biological activities3,4,5. Various quinolines have been reported as anti-malarial, anti-inflammatory, anti-asthmatic, anti-bacterial, anti-hypertensive and platelet derived growth factor receptor tyrosine kinase (PDGF-RTK)-inhibiting agents6. A large variety of quinolines are reported to exhibit substantial anti-cancer activity7,8,9,10,11,12,13,14 through a variety of mechanisms, such as cell cycle arrest in the G2 phase13, inhibition of topoisomerase15, inhibition of tubulin polymerization16 and the inhibition of tyrosine kinases17,18,19.
Herein, we are reporting the anti-cancer activity of ten benzo[h]quinoline derivatives against human skin cancer (G361), lung cancer (H460), breast cancer (MCF7) and colon cancer (HCT116) cell lines. The most potent compounds identified (3e, 3f, 3h and 3j) showed effective cytotoxicity against these cancer cell lines with IC50 values ranging from 4.7 to 7.6 μM. Molecular docking studies were also performed to predict the likely interactions of these molecules in the binding site of their human target receptors cyclin-dependent kinase-2 (CDK2) and aromatase. The oral bioavailability/drug-likeness of the compounds were evaluated through predictive absorption, distribution, metabolism, and excretion (ADME) screening. Oxidation of DNA (8-hydroxy-20-deoxyguanosine (8-OHdG) formation) extracted from all cancer cells was measured after exposure to active arylated benzo[h]quinolines. The action of these active compounds on oxidative signaling cascades was characterized by measuring the apoptosis-related mRNA gene expression of ATM (Ataxia telangiectasia mutated is a serine/threonine protein kinase)20, Bax (BCL2-associated X protein regulated by the tumor suppressor P53)21,22,23 and H2AFX (H2A histone family, member X)24. Apoptosis was studied by flow cytometry.
Results and Discussion
Chemistry
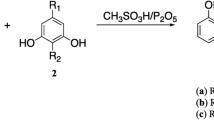
The synthesis of benzo[h]quinolines is presented in Fig. 1. The required precursor was synthesized in two steps. In the first step, synthesis of 6-aryl-4-methylthio-2-oxo-2H-pyran-3-carbonitriles was performed by the reaction of methyl 2-cyano-3,3-bis-methylthio-acrylate with various aryl/heteroaryl methyl ketones in dimethylsulphoxide (DMSO) under basic conditions at room temperature. Further, 6-aryl-4-sec.amino-2-oxo-2H-pyran-3-carbonitriles 1 was obtained by amination25 of 6-aryl-4-methylthio-2-oxo-2H-pyran-3-carbonitriles by various secondary amines in refluxing ethanol (Fig. 1)26,27,28. Synthesis of highly functionalized benzo[h]quinolines was carried out by stirring an equimolar mixture of 6-aryl-4-sec.amino-2H-pyran-2-one-3-carbonitriles, 2-cynomethylbenzonitrile 2 and sodium amide in N,N-dimethylformamide at 100 °C for 35–50 h (Fig. 1). The presence of an electron donating or withdrawing functional group on the aryl group at the C-6 position of the pyran ring did not affect the yield of the desired product. Interestingly, the presence of thienyl and furyl rings in lieu of an aryl ring required longer duration of the reaction and afforded good yields. Synthesis of 2-amino-5-aryl-4-sec.aminobenzo[h]quinoline-6-carbonitriles 3 containing piperidine and morpholine at the C-4 position was also carried out. Conventional heating was replaced with microwave-assisted heating to reduce the reaction time (Fig. 1). Heating of a mixture of 6-aryl-2-oxo-4-sec.amino-2H-pyran-3-carbonitriles 1, 2-cynomethylbenzonitrile and sodium amide in DMF at 100 °C for 55 minutes in a microwave reactor resulted excellent yields of 2-amino-5-aryl-4-sec.aminobenzo[h]quinoline-6-carbonitriles. All the studied compounds were synthesized by using the same procedure and characterized by spectroscopic analysis29,30,31.
Structure activity Relationship
On the basis of result obtained after evaluation of anticancer activity, we have tried to understand the role of different functional groups on the reactivity. As it is clear from Table 1 that benzo[h]quinolines (3e, 3f, 3h and 3j) are most active amongst reported compound. Compound 3f has an IC50 of 5.4, 4.7, 4.9 μM against H460, MCF7 and HCT116 cells respectively, which is close to the reference compound doxorubicin (2.1 μM) and similarly compound 3e exhibit IC50 value 5.3, 6.8, 7.6 and 6.8 against G361, H460, MCF7 and HCT116 cancer cell line respectively. From this result we conclude that compound with furyl group 3e and thienyl group 3f at position 5 of benzo[h]quinolines have good activity, which proposed that five member aromatic ring plays important role in activity. From the comparison of compound 3b and 3h it is clear that only change of piperidin to morpholine of benzo[h]quinolines affect the cytotoxicity in almost all of the cell lines, as shown in Table 1. The presence of the 4-methoxyphenyl group (3j) results in IC50values of 4.8, 5.2 and 6.8 μM against the H460, MCF7 and HCT116 cell lines, respectively, while the presence of the 4-bromophenyl group (3i) results in an IC50 value of 6.9 μM with the HCT116 cancer cell line. In summary, the presence of 4-chlorophenyl and 4-methoxyphenyl, furyl and theinyl ring in benzo[h]quinoline with suitable piperidine/morpholine moiety result in significant cytotoxicity.
Cytotoxicity and apoptosis studies
Compounds 3a–3j was evaluated for in vitro cytotoxic activity by MTT assay against four human cancer cell lines: G361, H460, MCF7 and HCT116. Doxorubicin was used as a positive control (Table 1). Results obtained reveals that 3f is cytotoxic against the HCT116 (IC50 4.9 μM), MCF7 (IC50 4.7 μM) and H460 (IC50 5.4 μM) cell lines shown in Fig. 2a–d. Another compound, 3e was more cytotoxic compared to doxorubicin against the G361 (IC50 5.3 μM), H460 and HCT116 (IC50 6.8 μM) cell lines. Compound 3h showed significant cytotoxicity against the G361 (IC50 5.5 μM), H460 (IC50 5.4 μM) and MCF7 (IC50 5.2 μM) cell lines but not against the HCT116 cell line. Apoptotic effect of active compound 3e was studied at its IC50 concentration (see Table 1) on G361, H460, MCF7 and HCT 116 cancer cells. In this assay, cells were harvested and annex in V-FITC/PI staining was performed to determine apoptotic activity. Compound 3e induced apoptosis in the range ~25–30% of cells in G361, H460, MCF7 and HCT116 cancer cell lines (Fig. 3). This result strongly suggests that activation of intracellular ROS and oxidative DNA damage might be responsible for induced apoptosis in these cancer cells.
Change in cell viability upon exposure to different concentrations of compounds 3e-3f in different cell lines: (a) G361; (b) H460 (c) MCF7 and (d) HCT116. In each experiment a negative control (no drug treatment) and a standard anticancer drug, Doxorubicin was used. All values are expressed as triplicate averages ± SD.
Intracellular ROS activation, mRNA expression and DNA oxidation analysis
Intracellular ROS levels were estimated using fluorescent probes. The ROS level detected by H2DCFDA staining was dramatically increased upon addition of compounds 3e, 3f, 3h and 3j in all the four cancer cell lines, and was significantly higher in comparison with the positive control (Fig. 4a,b). The increase in intracellular ROS may cause oxidative stress. Hence in order to investigate the effect of the above active compounds on DNA oxidation, formation of 8-oxoguanine as a typical oxidative base lesion 7,19,29,36 was measured through the increased formation of 8-OHdG, as shown in Fig. 4c. Exposure to all active compounds lead to a substantial increase in the amount of 8-OHdG in all cancer cells. This reveals that these compounds can activate oxidative stress signaling leading to DNA oxidation. Thus, this result suggested that 3e and 3f treatment markedly induced DNA damage in cancer cells. In Fig. 4d shows that compound 3e induce the cells death might be depends on inhibition of cyclin-dependent CDK2, a serine–threonine protein kinase associated with cell cycle progression and DNA damage. In order to establish the role of increased intracellular ROS in cancer cell death induced by the above active compounds, mRNA expression was evaluated for three oxidative stress-related genes, ATM, H2AX, and BAX (Fig. 5a–e). The ataxia telangiectasia mutated (ATM) is a serine/threonine protein kinase and responsible for activating cellular responses to DNA damage, which may be activated by H2AX and CDK2 in the DNA damage checkpoint32,33,34 but a mechanistic link between these two pathways has not been clearly elucidated. H2AX plays a key role in DNA damage response and is required for the assembly of DNA repair proteins at sites containing damaged chromatin as well as for activation of checkpoint proteins, which arrest the cell cycle progression35.
Changes in intracellular ROS and DNA oxidation.
(a) Level of intracellular ROS in all cancer cells after exposure tocompounds 3e, 3f, 3h, 3j and Doxorubicin. All values are expressed as the fluorescence intensity ratio between compound and control. All values are expressed as triplicate averages ± SD. (b) Qualitative analysis of intracellular ROS level after exposure to compound 3e in all cancer cells from fluorescence microscopy using the fluorescent probe H2DCFDA. (c) Amount of 8-OHdG production upon DNA oxidation upon exposure to compounds 3e, 3f, 3h, 3j and Doxorubicin. (d) Western blot analysis of CDK2 expression after exposure to compound 3e in all cancer cells. All values are expressed as triplicate averages ± SD. A Student t-test was performed with respect to the control (*denotes P < 0.05 and **denotes P < 0.01).
Apoptosis-related mRNA expression of H2AX, Bax, ATM, and 18 s rRNA (reference gene) after 24 h incubation with different compounds: (a) 3e, (b) 3f, (c) 3h (d) 3j and (e) Doxorubicin. The relative value of mRNA expression of these genes was measured by quantitative real-time PCR. All values are expressed as triplicate averages ± SD. A Student t-test was performed with respect to the control (* denotes P < 0.05 and ** denotes P < 0.01).
This study also demonstrates that exposure to active arylated benzo[h]quinolines increases levels of intracellular ROS, which may be partially mediated through changes in mitochondrial potential leading to cell death induced by H2AX and ATM over-expression. As shown in Fig. 5a–e reveals that cancer cells exposed to active compounds showed over expression of H2AX and ATM genes leading to DNA damage in cancer cells. Therefore, over expression of these genes plays a major role in initiating cell death in cancer cells through the DNA damage signalling pathway.
Molecular modeling
The synthesized benzo[h]quinolines were evaluated for their inhibitory effects in different cancer cell lines. Molecular docking studies were carried out to ascertain the mode of action towards the molecular targets, that is known human breast cancer target receptor aromatase (PDB ID: 3EQM)37,38 and colon cancer target protein CDK2 (PDB ID: 2R3J)39,40. Binding affinities were predicted by the Sybyl docking total score upon docking with the Surflex-Dock program (Sybyl X 2.0). Docking studies were carried out to evaluate the binding affinity and interactions with their target proteins. Hydrogen bonds (H-bonds, with a donor-receptor distance of 3 Å) between the ligand and amino acids in the binding site of the protein were used for the ranking of compounds. The mode of interaction of the co-crystallized ligand (3-bromo-5-phenyl-N-(pyridin-3-ylmethyl) pyrazolo [1,5- a]pyrimidin-7-amine) (SCJ) within the crystal structure of CDK2 was used as a reference binding model. The binding site of CDK2 contains the important residue Leu-83 is important for hydrophobic interactions with the ligand.
Doxorubicin (standard compound) was docked into CDK2 with a predicted total score of 5.1647. The hydrophobic cavity made by residues Asp-86, Lys-89 and Leu-83 interacted with the ligand through formation of H-bonds. Docking of compound 3e with CDK2 resulted in a predicted high binding affinity with a total score of 5.8273 and the formation of a strong H-bond (of length 2.0 Å) to the hydrophobic residue Leu-83 (Fig. 6). The predicted binding mode of compound 3e with CDK2 involved a number of binding site residues within a radius of 3 Å with diverse properties (Table 2):basic (polar, hydrophobic, positively charged), for example Lys-33, Lys-89, His-84; aromatic (hydrophobic), for example, Phe-82; acidic (polar, negatively charged), for example, Glu-12, Asp-145 and Asp-86; polar amide, for example, Gln-85, Gln-131 and Asn-132; hydrophobic, for example, Ala-177, Ala-131, Ala-144, Gly-13, Leu-83, Leu-134, Ile-10, Val-18 and Gly-11. As a consequence, compound 3e was predicted to have strong hydrophobic interactions with CDK2, which provides a molecular rational for the activity in this compound (Fig. 6). Docking of compound 3f also predicted high binding affinity with a total score of 5.5206 and the formation of a strong H-bond (length 2.1 Å) to the hydrophobic residue Leu-83, with interactions with almost the same amino acid residues in the binding site (Fig. 7).
Docking of compounds 3f, 3g, 3h and 3j with the breast cancer target protein aromatase (PDB ID:3EQM) was also carried out. For comparison, the docking score of the control anti-cancer drug doxorubicin was predicted to be 2.2253. Docking of compound 3 f resulted in a predicted high binding affinity with a total score of 5.4971 and the formation of an H-bond with Ala-438 (Table 3). The predicted binding mode of compound 3f with aromatase involved a number of binding site residues within a radius of 3 Å with diverse properties: hydrophobic, for example Met-374, Ala-438, Ala-306, Ile-133, Ile-132, Leu-372, Val-373, and Val-370; aromatic (hydrophobic), for example Phe-134; nucleophilic (polar, hydrophobic), for example Cys-437; nucleophilic (polar, hydrophobic), for example Thr-310, Cys-437; polar amide, for example Gln-86, Asn-105; hydrophobic positively charged, for example Arg-115; acidic (polar, negatively charged), for example, Asp-309. As a consequence, compound 3f was predicted to have strong hydrophobic interactions with aromatase, which provides a molecular rationale for its observed activity (Fig. 8).
Similarly, docking of compound 3h resulted in a predicted high binding affinity with a total score of 5.5392. The predicted binding mode of compound 3h with aromatase involved a number of binding site residues within a radius of 4 Å that were acidic (polar, negative charged), for example Asp-309. Compound 3 h was also predicted to have strong hydrophobic interactions with aromatase, which explain its observed activity (Fig. 9). Likewise, docking of compound 3j against aromatase gave a predicted high binding affinity with a total score of 5.0609 (Table 3). The predicted binding mode of compound 3j involved several binding site residues within a radius of 4 Å that were acidic (polar, negative charged), for example Asp-309, and hydrophobic, for example Met-374. The strong hydrophobic interactions involved in the binding of compound 3j explain its observed activity (Fig. 10).
Pharmacokinetic and toxicity properties
Pharmacokinetic (PK) ADME properties are important descriptors for human therapeutic use of any drug molecule. These ADME descriptors were calculated for all active molecules and compared with standard ranges. All the derivatives possessed a good number of hydrogen bond donors and acceptors39,40. These derivatives were designed to increase the binding of the drug with the receptor through hydrogen bonding. These derivatives were found to follow Lipinski’s rule of 5, affording drug likeness to the designed compounds. Polar surface area was calculated to estimate the ability of the compounds to permeate cell membranes. Lipophilicity (ratio of octanol solubility to water solubility) measured through logP, which has been implicated in blood brain barrier penetration and permeability prediction. Excretion of drugs depends on MW and logP41.
While the benzo[h]quinolines showed significant anti-cancer activity, they also exhibited some PK limitations. The calculated TPSA values of these compounds were within acceptable limits. The distribution of compounds in the human body was described by the predicted blood–brain barrier coefficient (logBB), apparent Caco-2 permeability, log Kp for skin permeability, volume of distribution and plasma protein binding (log Khsa for serum protein binding)42,43,44. All compounds show poor aqueous solubility (Table 1 in Supplementary Information). All compounds except 3g were predicted to be CYP2D6 non-inhibitors, and all derivatives were predicted to be hepatotoxic and highly-bound to plasma binding protein. Compounds 3c, 3d, 3i and 3j show medium apparent MDCK permeability. The calculated BBB values of the analogues were within the acceptable interval. The calculated values for these ADME parameters showed close similarity between the analogues and that of the reference drug doxorubicin and lie within the standard range of values exhibited by 95% of all known drugs shown in Table 4 and supplementary Figure S1.
Toxicity risks assessment
The reproductive/developmental toxicity, Ames mutagenicity, skin irritant effects, Ames score, rat oral LD50 (mg/kg), and rat carcinogenic potency TD50 (mg/kg) were predicted for all benzo[h]quinoline derivatives. These compounds were estimated to possess no risk of skin irritation. Compounds 3c, 3d, 3i and 3j may show developmental toxicity. All compounds were predicted to possess high mutagenicity potential at high doses or long term therapeutic use in human, except for compound 3c, which was predicted to be non-mutagenic45,46. Other predicted toxicity parameters are summarized in Table 5. The toxicity risks for benzo[h]quinoline derivative scanned to find the moderate to good compared with doxorubicin. Similarly, toxicity screening for USFDA rodent carcinogenicity, Ames mutagenicity, developmental toxicity potential, and carcinogenic effect irritancy are predicted to have a positive response for these benzo[h]quinoline derivatives.
Material Methods
Chemical synthesis
General experimental procedures
All the reagents and solvents used in this study were purchased from Sigma Aldrich and Alfa Aesar. All compounds were synthesized by the method described by Singh et. al.20 (Detail data available in S1). Detailed structural characterization is mentioned therein. These derivatives were defined as benzo[h]quinoline anti-cancer derivatives. In this study, we have further explored their cytotoxic effect on G361, H460, MCF7 and HCT116 cancer cell lines using in-vitro, and in-silico bioassays.
Reagents and consumables
G361, H460, MCF7 and HCT116 cancer cell lines were procured from KCLB (Korean Cell Line Bank, South Korea). All the cells were maintained in RPMI and DMEM supplemented with 10% fetal bovine serum, 1% non-essential amino acids, 1% L-glutamine (300 μg/mL), 1% penicillin (100 IU/ml) and streptomycin (100 mg/ml) (Hyclone, USA)32,36,47. Cultured cells were grown at 37 °C, 95% relative humidity and 5% CO2 and passaged twice a week. Cells were allowed to grow in 75 cm2 tissue culture flasks until confluence and then sub-cultured for experimentation.
Cell viability assays by MTT
Cell viability was measured by the MTT assay34,36,48. Briefly, 2 × 105 cells/well were seeded in 96-well plates and incubated for 12 h at 37 °C in a 5% CO2 atmosphere to allow cell adhesion. Stock solutions of the compounds made in DMSO were sterilized through 0.45 micro size filter (Sartorus Stedim, biotech, USA), then further diluted to 0.2 mg/ml in incomplete medium for treatment against G361, H460, MCF7 and HCT116 cancer cells lines. A 100 μL solution of compound (3e, 3f, 3h and 3j) was added respectively to a 100-μL of fresh medium in wells to give final concentrations of 5–0.312 μM/mL after serial dilution. A negative control (without drug), a positive control with anticancer drug doxorubicin was included in each assay34,36.
Intracellular reactive oxygen species analysis
Total ROS content inside the cells was determined using the H2DCFDA reagent. A total of 2 × 105 cells/ml of G361, H460, MCF7 and HCT116 cancer cells in DMEM was incubated at the IC50 concentrations of the active compounds 3e, 3f, 3h, 3j and the positive control doxorubicin respectively and then transferred to a micro-centrifuge tube. The cells were washed with PBS and 500 μl of 10 mM H2DCFDA was added. After incubation at 30 °C for 1 h, the cells were washed twice with PBS. The cells were then recovered in PBS at 30 °C for 30 min and read at 495/515 (ex./em.) nm using a microplate reader34,36,47. The ratio of fluorescence intensity (FI = fluorescent value at excitation wavelength of 485 nm and an emission at 535 nm of treated cells/control) was calculated between exposure and control (no treated) cells.
RNA extraction for quantitative real time PCR
We measured mRNA expression of three apoptosis related genes, including H2AX, ATM, and BAX. The total cellular RNA was isolated from all four cancer cells line after 24 h incubated at the IC50 concentration of 3e, 3f, 3h, 3j and Doxorubicin respectively. Primer design, cDNA synthesis and quantification of every gene was carried out as previously reported34,36. Quantitative real-time PCR was done by using following forward and reverse primer sequences, as listed below. 18sRNA was amplified to ensure cDNA integrity and to normalize expression.
Primer sequences used for mRNA expression.

Apoptosis analysis by annexin V-FITC/PI staining
Cancer cells were exposed with IC50 concentration of active compound 3e to detect apoptosis using the annexin V-FITC apoptosis detection kit. Treated/untreated cells were trypsinized after incubation for 12 h, after first washing with 1 ml of cold 1× biding buffer. Annexin V-FITC (0.5 mg ml−1) was added to each sample. After incubation for 15 min at room temperature, the cells were again washed with PBS and stained with 0.3 mg ml−1 of PI (Propidium Iodide) and analyzed by flow cytometry (BD FACS Verse, BD Biosciences)34,36,47.
DNA damage analysis
Genomic DNA was extracted from all four cell lines after treatment with IC50 concentrations of 3e, 3f, 3h, 3j and Doxorubicin respectively, for OH-dG detection. Genomic DNA was extracted from treated/untreated cells following a standard molecular biology protocol and re-suspended in 50 μl water34,36. The same amount of genomic DNA (2 mg) extracted from cells was used for the detection of 8-OH-dG level following the standard protocol of anoxidative DNA damage ELISA kit (Cell Biolabs, Inc. USA)34,36.
Western blot analysis
All cells were exposed with IC50 concentrations of 3e compounds. After exposure, protein extraction of the cells was performed. The whole cell protein extracts from treated/untreated cells were lysed in a RIPA buffer (Cell Signaling Technology, USA) and the extracted proteins were subjected to electrophoresis in 12% SDS-PAGE and blotted onto nitrocellulose membranes36,47,48,49. The membrane was probed with the CDK2 antibodies for protein expression, (Cell Signaling Technology, USA)50,51,52. The bands were detected using the Super Signal West Pico Chemiluminescent substrate (Pierce, Rockford, IL, USA) and images was taken using a Vilver imaging system (Vilver, Upland, CA, USA).
Molecular docking study
Molecular modeling studies of benzo[h]quinoline derivatives were carried out using molecular modeling software Sybyl-X 2.0, (Tripos International, St. Louis, Missouri, 63144, USA). Drawing of structures and simple geometry optimization were performed with Chem Bio-Office suite Ultra v12.0 (2012) (Cambridge Soft Corp., UK). Docking of all compounds was carried out on the human anticancer targets aromatase (PDB ID: 3EQM)37,38 and CDK2 (PDB ID: 2R3J)39,40. The Surflexdoc module in Sybyl was used to construct a 3D model of the structures53,54. Details data point available in S1 of supplementary material.
Screening through in silico pharmacokinetic parameters
Pharmacokinetic (PK) properties depend on chemical descriptors of drugs, which determine their absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties, which are the key descriptors for the human therapeutic use of any compound. These PK parameters were calculated using the ADMET modules in Discovery Studio v3.5 software (Accelrys, USA), and are reported in supplementary Table S1 of the Supplementary Information. Predictive mathematical ADMET models were derived with different PK parameters, namely aqueous solubility, blood-brain barrier penetration, cytochrome P-450 2D6 inhibition, hepatotoxicity, human intestinal absorption and plasma protein binding. Predictions from these models were used to quantitatively predict properties of a set contrasted with of known rules that specify for appropriate ADMET characteristics of the chemical structure of all benzo[h]quinoline derivatives.
Statistical analysis
All values are presented as the mean ± SD of the indicated number of replicates. Statistical analyses of the data were performed using Student’s t-test, and significant differences were based on P < 0.05 or P < 0.0134,36,48.
Additional Information
How to cite this article: Yadav, D. K. et al. New arylated benzo[h]quinolines induce anti-cancer activity by Oxidative stress-mediated DNA damage. Sci. Rep. 6, 38128; doi: 10.1038/srep38128 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
WHO. World Cancer Report. World Health Organization. 9283204298, 630 (2014).
Ferreira, L. R. et al. In vitro Cytotoxic Activity of β-chalcogen-substituted Michael-aldol Type Adducts Against Hela and RKO Cell Lines. Journal of Biological Sciences 13, 628–633, doi: 10.3923/jbs.2013.628.633 (2013).
Larsen, R. D. et al. Practical Route to a New Class of LTD4 Receptor Antagonists. The Journal of Organic Chemistry 61, 3398–3405 (1996).
Roma, G., Di Braccio, M., Grossi, G., Mattioli, F. & Ghia, M. 1,8-Naphthyridines IV. 9-substituted N,N-dialkyl-5-(alkylamino or cycloalkylamino) [1,2,4]triazolo[4,3-a][1, 8]naphthyridine-6-carboxamides, new compounds with anti-aggressive and potent anti-inflammatory activities. European Journal of Medicinal Chemistry 35, 1021–1035 (2000).
Chen, Y. L., Fang, K. C., Sheu, J. Y., Hsu, S. L. & Tzeng, C. C. Synthesis and antibacterial evaluation of certain quinolone derivatives. Journal of Medicinal Chemistry 44, 2374–2377 (2001).
Dubé, D. et al. Quinolines as potent 5-lipoxygenase inhibitors: Synthesis and biological profile of L-746,530. Bioorganic & Medicinal Chemistry Letters 8, 1255–1260, doi: 10.1016/S0960-894X(98)00201-7 (1998).
Alqasoumi, S. I., Al-Taweel, A. M., Alafeefy, A. M., Noaman, E. & Ghorab, M. M. Novel quinolines and pyrimido[4,5-b]quinolines bearing biologically active sulfonamide moiety as a new class of antitumor agents. European Journal of Medicinal Chemistry 45, 738–744, doi: 10.1016/j.ejmech.2009.11.021 (2010).
Behforouz, M. et al. Synthesis and evaluation of antitumor activity of novel N-acyllavendamycin analogues and quinoline-5,8-diones. Bioorganic & medicinal chemistry 15, 495–510, doi: 10.1016/j.bmc.2006.09.039 (2007).
Kemnitzer, W. et al. Discovery of 1-benzoyl-3-cyanopyrrolo[1,2-a]quinolines as a new series of apoptosis inducers using a cell- and caspase-based high-throughput screening assay. Part 1: Structure-activity relationships of the 1- and 3-positions. Bioorganic & Medicinal Chemistry Letters 18, 6259–6264, doi: 10.1016/j.bmcl.2008.09.110 (2008).
Ferlin, M. G., Gatto, B., Chiarelotto, G. & Palumbo, M. Novel pyrrolo[3,2-f]quinolines: synthesis and antiproliferative activity. Bioorganic & Medicinal chemistry 9, 1843–1848 (2001).
Abouzid, K. & Shouman, S. Design, synthesis and in vitro antitumor activity of 4-aminoquinoline and 4-aminoquinazoline derivatives targeting EGFR tyrosine kinase. Bioorganic & Medicinal Chemistry 16, 7543–7551, doi: 10.1016/j.bmc.2008.07.038 (2008).
Gopal, M., Shenoy, S. & Doddamani, L. S. Antitumor activity of 4-amino and 8-methyl-4-(3diethylamino propylamino)pyrimido[4′,5′:4,5]thieno (2,3-b) quinolines. Journal of Photochemistry and photobiology. B, Biology 72, 69–78 (2003).
Kim, Y. H. et al. G2 arrest and apoptosis by 2-amino-N-quinoline-8-yl-benzenesulfonamide (QBS), a novel cytotoxic compound. Biochemical Pharmacology 69, 1333–1341, doi: 10.1016/j.bcp.2004.12.019 (2005).
Zhao, Y. L., Chen, Y. L., Chang, F. S. & Tzeng, C. C. Synthesis and cytotoxic evaluation of certain 4-anilino-2-phenylquinoline derivatives. European Journal of Medicinal Chemistry 40, 792–797, doi: 10.1016/j.ejmech.2005.03.008 (2005).
Cheng, Y. et al. Synthesis, cytotoxic activities and structure-activity relationships of topoisomerase I inhibitors: indolizinoquinoline-5,12-dione derivatives. Bioorganic & Medicinal Chemistry 16, 4617–4625, doi: 10.1016/j.bmc.2008.02.036 (2008).
Alqasoumi, S. I. et al. Synthesis and biological evaluation of 2-amino-7,7-dimethyl 4-substituted-5-oxo-1-(3,4,5-trimethoxy)-1,4,5,6,7,8-hexahydro-quinoline-3-carbon itrile derivatives as potential cytotoxic agents. Bioorganic & Medicinal Chemistry Letters 19, 6939–6942, doi: 10.1016/j.bmcl.2009.10.065 (2009).
Mulvihill, M. J. et al. Novel 2-phenylquinolin-7-yl-derived imidazo[1,5-a]pyrazines as potent insulin-like growth factor-I receptor (IGF-IR) inhibitors. Bioorganic & Medicinal Chemistry 16, 1359–1375, doi: 10.1016/j.bmc.2007.10.061 (2008).
Nishii, H. et al. Discovery of 6-benzyloxyquinolines as c-Met selective kinase inhibitors. Bioorganic & Medicinal Chemistry Letters 20, 1405–1409, doi: 10.1016/j.bmcl.2009.12.109 (2010).
Pannala, M. et al. Synthesis and structure-activity relationship of 4-(2-aryl-cyclopropylamino)-quinoline-3-carbonitriles as EGFR tyrosine kinase inhibitors. Bioorganic & Medicinal Chemistry Letters 17, 5978–5982, doi: 10.1016/j.bmcl.2007.07.071 (2007).
Ahmed, M. & Rahman, N. ATM and breast cancer susceptibility. Oncogene 25, 5906–5911, doi: 10.1038/sj.onc.1209873 (2006).
Miyashita, T. et al. Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene 9, 1799–1805 (1994).
Selvakumaran, M. et al. Immediate early up-regulation of bax expression by p53 but not TGF beta 1: a paradigm for distinct apoptotic pathways. Oncogene. 1994 Jun;9(6):1791-8. 9, 1791–1798 (1994).
Toshiyuki, M. & Reed, J. C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 80, 293–299, doi: 10.1016/0092-8674(95)90412-3 (1995).
Sharma, A., Singh, K. & Almasan, A. Histone H2AX phosphorylation: a marker for DNA damage. Methods Mol Biol 920, 613–626, doi: 10.1007/978-1-61779-998-3_40 (2012).
Tominaga, Y., Ushirogochi, A. & Matsuda, Y. Synthesis and reaction of 6-substituted 3-methoxycarbonyl-4-methylthio-2H-pyran-2-one derivatives. Journal of Heterocyclic Chemistry 24, 1557–1567, doi: 10.1002/jhet.5570240612 (1987).
Pratap, R., Kumar, B. & Ram, V. J. Substituent-induced regioselective synthesis of 1,2-teraryls and pyrano[3,4-c]pyran-4,5-diones from 2H-pyran-2-ones. Tetrahedron 62, 8158–8163 (2006).
Farhanullah, F., Agarwal, N., Goel, A. & Ram, V. J. Synthesis of aminonicotinonitriles and diaminopyridines through base-catalyzed ring transformation of 2H-pyran-2-ones. The Journal of Organic Chemistry 68, 2983–2985, doi: 10.1021/jo0204496 (2003).
Pratap, R. et al. Molecular Docking and Biological Evaluation of Functionalized benzo[h]quinolines as Colon Cancer Agents. Bioinformatics and Biomedical Engineering 9044, 664–673, doi: 10.1007/978-3-319-16480-9_64 (2015).
Singh, S. et al. One-Pot Chemoselective Synthesis of Arylated Benzo[h]quinolines. Synlett 25, 2599–2604 (2014).
Singh, S. et al. Base mediated regioselective synthesis of highly functionalized conjugated enones. Tetrahedron Letters 56, 5203–5208 (2015),
Singh, S. et al. Precursor directed regioselective synthesis of partially reduced benzo[e]indene through oxidative cyclization and benzo[h]quinolines. RSC Advances 5, 18335–18341 (2015).
Bedard, K. & Krause, K. H. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiological Reviews 87, 245–313, doi: 10.1152/physrev.00044.2005 (2007).
Yamaura, M. et al. NADPH oxidase 4 contributes to transformation phenotype of melanoma cells by regulating G2-M cell cycle progression. Cancer Research 69, 2647–2654, doi: 10.1158/0008-5472.CAN-08-3745 (2009).
Kumar, N. et al. The action of microsecond-pulsed plasma-activated media on the inactivation of human lung cancer cells. Journal of Physics D: Applied Physics 49, 115401 (2016).
Podhorecka, M., Skladanowski, A. & Bozko, P. H2AX Phosphorylation: Its Role in DNA Damage Response and Cancer Therapy. Journal of Nucleic Acids 2010, doi: 10.4061/2010/920161 (2010).
Kumar, N. et al. Induced apoptosis in melanocytes cancer cell and oxidation in biomolecules through deuterium oxide generated from atmospheric pressure non-thermal plasma jet. Scientific Reports 4, 7589, doi: 10.1038/srep07589 (2014).
Suvannang, N., Nantasenamat, C., Isarankura-Na-Ayudhya, C. & Prachayasittikul, B. Molecular Docking of Aromatase Inhibitors. Molecules 16, 3597–3617, doi: 3390/molecules16053597 (2011).
Ghosh, D., Griswold, J., Erman, M. & Pangborn, W. Structural basis for androgen specificity and oestrogen synthesis in human aromatase. Nature 457, 219–223, doi: 10.1038/nature07614 (2009).
Yoon, H. et al. Design, synthesis and inhibitory activities of naringenin derivatives on human colon cancer cells. Bioorganic & Medicinal Chemistry Letters 23, 232–238, doi: 10.1016/j.bmcl.2012.10.130 (2013).
Kelly, M. D. & Mancera, R. L. Comparative analysis of the surface interaction properties of the binding sites of CDK2, CDK4, and ERK2. ChemMedChem 1, 366–375, doi: 10.1002/cmdc.200500033 (2006).
Yadav, D. K. et al. QSAR and docking studies on chalcone derivatives for antitubercular activity against M. tuberculosis H37Rv. Journal of Chemometrics 28, 499–507 (2014).
Yadav, D. K. & Khan, F. QSAR, docking and ADMET studies of camptothecin derivatives as inhibitors of DNA topoisomerase-I. Journal of Chemometrics 27, 21–23 (2013).
Yadav, D. K. et al. Molecular docking and ADME studies of natural compounds of Agarwood oil for topical anti-inflammatory activity. Current Computer-Aided Drug Design 9, 360–370 (2013).
Yadav, D. K., Kalani, K., Khan, F. & Srivastava, S. K. QSAR and docking based semi-synthesis and in vitro evaluation of 18 beta-glycyrrhetinic acid derivatives against human lung cancer cell line A-549. Med Chem 9, 1073–1084 (2013).
Yadav, D. K., Khan, F. & Negi, A. S. Pharmacophore modeling, molecular docking, QSAR, and in silico ADMET studies of gallic acid derivatives for immunomodulatory activity. Journal of Molecular Modeling 18, 2513–2525, doi: 10.1007/s00894-011-1265-3 (2012).
Yadav, D. K. et al. Development of QSAR model for immunomodulatory activity of natural coumarinolignoids. Drug design, development and therapy 4, 173–186 (2010).
Kumar, N. Influence of water vapour with non-thermal plasma jet on the apoptosis of SK-BR-3 breast cancer cells. RSC Advances 5, 14670–14677 (2015).
Attri, P. et al. Influence of reactive species on the modification of biomolecules generated from the soft plasma. Scientific Reports 5, 8221, doi: 10.1038/srep08221 (2015).
Jackson, S. P. DNA damage detection by DNA dependent protein kinase and related enzymes. Canc. Surv. 28, 261–279 (1996).
Durocher, D. & Jackson, S. P. DNA-PK, ATM and ATR as sensors of DNA damage: variations on a theme? Curr. Opin. Cell. Biol. 13, 225–231 (2001).
Yang, J., Yu, Y., Hamrick, H. E. & Duerksen-Hughes, P. J. ATM, ATR and DNA-PK: initiators of the cellular genotoxic stress responses. Carcinog. 24, 1571–1580 (2003).
Shiloh, Y. & Ziv, Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell. Biol. 14, 197–210 (2013).
Yadav, D. K. et al. QSAR and docking based semi-synthesis and in vivo evaluation of artemisinin derivatives for antimalarial activity. Current Drug Targets 15, 753–761 (2014).
Yadav, D. K. et al. Design, synthesis and in vitro evaluation of 18beta-glycyrrhetinic acid derivatives for anticancer activity against human breast cancer cell line MCF-7. Current Medicinal Chemistry 21, 1160–1170 (2014).
Acknowledgements
The authors thank the Council of Scientific and Industrial Research (CSIR, New Delhi) and DST, New Delhi [DST-DU Purse grant] for financial support. RR Thanks to DST, New Delhi for Women Scientist Fellowship. The authors thank the University of Delhi for providing research funding and access to instrumentation facility. DKY acknowledges the Science & Engineering Research Board, New Delhi for financial support through the Young Scientist Project SB/YS/LS-130/2014 at the All India Institute of Medical Sciences (AIIMS), Jodhpur, India and National Research Foundation (NRF) of Korea, which is funded (No: 2012R1A6A3A04038302) by the Ministry of Education, Science, and Technology, Korea.
Author information
Authors and Affiliations
Contributions
R.P. and D.K.Y. conceived and designed the experiments, analyzed the data and wrote the manuscript. N.K., P.S.,R.R. D.K.Y. and P.S. performed in vitro research work. D.K.Y. carried out all molecular modeling work. R.P. and S.S. designed and synthesized the compounds. H.P.S., M.H.K., S.M., R.L.M., E.H.C. and P. S. provided the molecular modeling lab facility. R.P., R.R. and D.K.Y. conceived the study, and participated in coordination of work and editing the manuscript. All authors read and approved the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Yadav, D., Rai, R., Kumar, N. et al. New arylated benzo[h]quinolines induce anti-cancer activity by oxidative stress-mediated DNA damage. Sci Rep 6, 38128 (2016). https://doi.org/10.1038/srep38128
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep38128
This article is cited by
-
Anticancer Activity–Structure Relationship of Quinolinone-Core Compounds: An Overall Review
Pharmaceutical Chemistry Journal (2023)
-
Cold atmospheric plasma generated reactive species aided inhibitory effects on human melanoma cells: an in vitro and in silico study
Scientific Reports (2020)
-
Reactions of quinoline-2(6,8)-carbaldehydes with arenes by the action of various Brønsted or Lewis acids: synthesis of diarylmethylquinolines
Chemistry of Heterocyclic Compounds (2020)
-
Molecular Insights into the Interaction of RONS and Thieno[3,2-c]pyran Analogs with SIRT6/COX-2: A Molecular Dynamics Study
Scientific Reports (2018)
-
Synthesis, docking and anticancer activity of azo-linked hybrids of 1,3,4-thia-/oxadiazoles with cyclic imides
Molecular Diversity (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.